- Mission, Facts and Figures
- Deans, Chairs and Staff
- Leadership Council
- Dean in the News
- Get Involved
- DEIB Mission
- Message from DEIB Associate Dean
- News and Media
- Reading Lists
- The Yale and Slavery Research Project
- Photo Gallery
- Winslow Medal
- Coat of Arms & Mace
- $50 Million Challenge
- For Pandemic Prevention and Global Health
- For Understanding the Health Impacts of Climate Change
- For Health Equity and Justice
- For Powering Health Solutions through Data Science
- For Future Leaders
- For Faculty Leaders
- For Transformational Efforts
- Data, Leadership, and Collaboration at the School of Public Health
- An abiding love for Yale turns into a lasting gift – in 15 minutes
- Endowed Professorship Created at Critical Time for Yale School of Public Health
- Brotherly encouragement spurs gift to support students
- Prestipino creates opportunities for YSPH students, now and later
- Alumna gives back to the school that “opened doors” in male-dominated field
- For Public Health, a Broad Mission and a Way to Amplify Impact
- Couple Endows Scholarship to Put Dreams in Reach for YSPH Students
- A Match Made at YSPH
- A HAPPY Meeting of Public Health and the Arts
- Generous Gift Bolsters Diversity & Inclusion
- Alumni Donations Aid Record Number of YSPH Students
- YSPH’s Rapid Response Fund Needs Donations – Rapidly
- Podiatric Medicine and Orthopedics as Public Health Prevention
- Investing in Future Public Health Leaders
- Support for Veterans and Midcareer Students
- Donor Eases Burden for Policy Students
- A Personal Inspiration for Support of Cancer Research
- Reducing the Burden of Student Debt
- Learning About Global Health Through Global Travel
- A Meeting in Dubai, and a Donation to the School
- Rapid Response Fund
- Planned Giving
- Testimonials
- Assistant/Associate Professor - Chronic Disease Epidemiology
- Assistant/Associate Professor - Environmental Toxicology
- Associate Research Scientist - Health Policy and Management
- Postdoctoral Associate - Computational Biology
- LGBTQ Mental Health Postdoctoral Clinical Research Associate in NYC
- Postdoctoral Associate - Health Policy and Management
- Postdoctoral Associate - Data Science and Data Equity
- Postdoctoral Associate - CMIPS
- Postdoctoral Associate - Environmental Health Sciences
- Postdoctoral Associate - Social and Behavioral Sciences
- Postdoctoral Associate - Malaria Genomics and Vaccinology
- Postdoctoral Associate - McDougal Lab
- Postgraduate Associate - Data, Modeling, and Decision Analysis
- For the Media
- Issues List
- PDF Issues for Download
- Editorial Style Guide
- Social Media
- Shared Humanity Podcast
- Health & Veritas Podcast
- Maps and Directions
- Accreditation
- Faculty Directory by Name
- Career Achievement Awards
- Annual Research Awards
- Teaching Spotlights
- Biostatistics
- Chronic Disease Epidemiology
- Climate Change and Health Concentration
- Environmental Health Sciences
- Epidemiology of Microbial Diseases
- Global Health
- Health Policy and Management
- Implementation Science Track
- Maternal and Child Health Promotion Track
- Public Health Modeling Concentration
- Social & Behavioral Sciences
- U.S. Health Justice Concentration
- Why Public Health at Yale
- Events and Contact
- What Does it Take to be a Successful YSPH Student?
- How to Apply and FAQs
- Orientation Schedule
- Traveling to Yale
- Meet Students and Alumni
- Past Internship Spotlights
- Student-run Organizations
- MS and PhD Student Leaders
- Staff Spotlights
- Life in New Haven
- Libraries at Yale
- The MPH Internship Experience
- Practicum Course Offerings
- Summer Funding and Fellowships
- Downs Fellowship Committee
- Stolwijk Fellowship
- Climate Change and Health
- Career Management Center
- What You Can Do with a Yale MPH
- MPH Career Outcomes
- MS Career Outcomes
- PhD Career Outcomes
- Employer Recruiting
- Tuition and Expenses
- External Funding and Scholarships
- External Fellowships for PhD Candidates
- Alumni Spotlights
- Bulldog Perks
- Stay Involved
- Update Your Info
- Board of Directors
- Emerging Majority Affairs Committee
- Award Nomination Form
- Board Nomination Form
- Alumni Engagement Plus
- Mentorship Program
- The Mentoring Process
- For Mentors
- For Students
- Recent Graduate Program
- Transcript and Verification Requests
- Applied Practice and Student Research
- Competencies and Career Paths
- Applied Practice and Internships
- Student Research
- Seminar and Events
- Competencies and Career paths
- Why the YSPH Executive MPH
- Message from the Program Director
- Two-year Hybrid MPH Schedule
- The Faculty
- Student Profiles
- Newsletter Articles
- Approved Electives
- Physicians Associates Program
- Joint Degrees with International Partners
- MS in Biostatistics Standard Pathway
- MS Implementation and Prevention Science Methods Pathway
- MS Data Sciences Pathway
- Internships and Student Research
- Competencies
- Degree Requirements - Quantitative Specialization
- Degree Requirements - Clinical Specialization
- Degree Requirements- PhD Biostatistics Standard Pathway
- Degree Requirements- PhD Biostatistics Implementation and Prevention Science Methods Pathway
- Meet PhD Students in Biostatistics
- Meet PhD Students in CDE
- Degree Requirements and Timeline
- Meet PhD Students in EHS
- Meet PhD Students in EMD
- Meet PhD Students in HPM
- Degree Requirements - PhD in Social and Behavioral Sciences
- Degree Requirements - PhD SBS Program Maternal and Child Health Promotion
- Meet PhD Students in SBS
- Differences between MPH and MS degrees
- Academic Calendar
- Translational Alcohol Research Program
- Molecular Virology/Epidemiology Training Program (MoVE-Kaz)
- For Public Health Practitioners and Workforce Development
- Course Description
- Instructors
- Registration
- Coursera Offerings
- Non-degree Students
- International Initiatives & Partnerships
- NIH-funded Summer Research Experience in Environmental Health (SREEH)
- Summer International Program in Environmental Health Sciences (SIPEHS)
- 2023 Student Awards
- 2022 Student Awards
- APHA Annual Meeting & Expo
- National Public Health Week (NPHW)
- Leaders in Public Health
- YSPH Dean's Lectures
- The Role of Data in Public Health Equity & Innovation Conference
- Innovating for the Public Good
- Practice- and community-based research and initiatives
- Practice and community-based research and initiatives
- Activist in Residence Program
- The Data & The Solutions
- Publications
- Health Care Systems and Policy
- Heart Disease and Stroke
- Panels, Seminars and Workshops (Recordings)
- Rapid Response Fund Projects
- SalivaDirect™
- Emerging Infections Program - COVID-NET
- Public Health Modeling Unit Projects
- HIV-AIDS-TB
- The Lancet 2023 Series on Breastfeeding
- 'Omics
- News in Biostatistics
- Biostatistics Overview
- Seminars and Events
- Seminar Recordings
- Statistical Genetics/Genomics, Spatial Statistics and Modeling
- Causal Inference, Observational Studies and Implementation Science Methodology
- Health Informatics, Data Science and Reproducibility
- Clinical Trials and Outcomes
- Machine Learning and High Dimensional Data Analysis
- News in CDE
- Nutrition, Diabetes, Obesity
- Maternal and Child Health
- Outcomes Research
- Health Disparities
- Women's Health
- News in EHS
- EHS Seminar Recordings
- Climate change and energy impacts on health
- Developmental origins of health and disease
- Environmental justice and health disparities
- Enviromental related health outcomes
- Green chemistry solutions
- Novel approaches to assess environmental exposures and early markers of effect
- 1,4 Dioxane
- Reproducibility
- Tissue Imaging Mass Spectrometry
- Alcohol and Cancer
- Olive Oil and Health
- Lightning Talks
- News in EMD
- Antimicrobial Resistance
- Applied Public Health and Implementation Science
- Emerging Infections and Climate Change
- Global Health/Tropical Diseases
- HIV and Sexually Transmitted Infections
- Marginalized Population Health & Equity
- Pathogen Genomics, Diagnostics, and Molecular Epidemiology
- Vector-borne and Zoonotic Diseases
- Disease Areas
- EMD Research Day
- News in HPM
- Health Systems Reform
- Quality, Efficiency and Equity of Healthcare
- Substance Abuse and Mental Health
- Modeling: Policy, Operations and Disease
- Pharmaceuticals, Vaccines and Medical Devices
- Health and Wellbeing
- News in SBS
- Aging Health
- Community Engagement
- Health Equity
- Mental Health
- Reproductive Health
- Sexuality and Health
- Nutrition, Exercise
- Stigma Prevention
- Community Partners
- For Public Health Practitioners
- Reports and Publications
- Fellows Stipend Application
- Agency Application
- Past Fellows
- PHFP in the News
- Frequently Asked Questions
- International Activity
- Research Publications
- Grant Listings
- Modeling Analyses
- 3 Essential Questions Series
INFORMATION FOR
- Prospective Students
- Incoming Students
- myYSPH Members

Doctor of Philosophy
The primary mission of the PhD program is to provide scholars with the disciplinary background and skills required to contribute to the development of our understanding of better ways of measuring, maintaining, and improving the public’s health. Examples of research conducted by PhD students includes but is not limited to: cancer epidemiology, clinical trials, cardiovascular disease, molecular epidemiology, vector-borne diseases, parasitology, mental health epidemiology and HIV/AIDS. Students are encouraged to work with faculty throughout the university since much of the work done in EPH is interdisciplinary.
How to Apply
Applications are submitted through the Graduate School of Arts and Sciences .
Select program: "Public Health" and your Concentration: Biostatistics (PhD or MS), Chronic Disease Epidemiology (PhD or MS), Environmental Health Sciences (PhD), Epidemiology of Microbial Diseases (PhD) or Epidemiology Infectious Disease (MS), Health Informatics (MS) Health Policy and Management (PhD) or Social and Behavioral Sciences (PhD).
The GRE and TOEFL code for Yale GSAS is: 3987. A writing sample is not required.
The deadline is December 15th.
PhD Program
All PhD students are guaranteed five years of 12-month stipend and tuition support in the form of YSPH fellowships, teaching fellowships, traineeships and research assistantships. In addition to support for tuition and living costs, students receive a health award to covers the full cost of single-student Yale Health Plan Hospitalization/Specialty Coverage.
Faculty Advisors
PhD applicants are not required to secure a faculty mentor prior to applying to the program.
We expect applicants to provide information in their personal statement about the research they hope to conduct if admitted and to state the faculty in our department whose research aligns with their interests.
Diversity Research Awards
The PhD program in Public Health enhances commitment its PhD students who identify as underrepresented minority students, first-generation college graduates and students from economically disadvantaged backgrounds by offering research awards to the top candidates admitted to the program. Each year a minimum of two PhD admitted students will be offered $2,000 each for research funds in addition to their financial aid package. Recipients have up to 2 years to spend these funds, which can be used for books, computers, software, conference travel, research travel or research supplies.
This funding is offered upon acceptance into the program. The criteria for the award is:
- Previous involvement in diversity-related initiatives in their community and/or volunteer activities helping underserved populations.
- Research interest in serving an underserved population
External Fellowships
Doctor of philosophy (phd) overview.
- See us on twitter
- See us on facebook
- See us on youtube
PhD in Epidemiology and Clinical Research
The PhD program in epidemiology and clinical research provides methodologic and interdisciplinary training to equip students to carry out cutting-edge epidemiologic research. The program trains students in the tools of modern epidemiology, with heavy emphasis on statistics, computer science, genetics, genomics, and bioinformatics. We welcome applicants with diverse backgrounds.
Pre-Application Sessions and Recordings

We are EPH: Meet Sam Jaros
Sam Jaros is a fourth year PhD candidate in the Department of Epidemiology and Population Health. His current research and thesis projects are focused on finding actionable patterns in opioid addiction to better spend limited public health resources on improving care. Sam developed a passion for improving care for opioid addiction while working in Appalachia in previous mining towns.
Read the Q&A with Sam

Student Bios

The Department of Epidemiology and Population Health is committed to fostering a diverse community in which all individuals are welcomed, respected, and supported to achieve their full potential.
Stanford recognizes that the Supreme Court issued a ruling in June 2023 about the consideration of certain types of demographic information as part of an admission review. All applications submitted during upcoming application cycles will be reviewed in conformance with that decision.
The Department of Epidemiogy and Population Health welcomes graduate applications from individuals with a broad range of life experiences, perspectives, and backgrounds who would contribute to our community of scholars. The review process is holistic and individualized, considering each applicant’s academic record and accomplishments, letters of recommendation, prior research experience, and admissions essays to understand how an applicant’s life experiences have shaped their past and potential contributions to their field and how they might enrich the learning community at Stanford.
Methods for Policy Research
Political analysis.
Boston University Academics
Boston University
- Campus Life
- Schools & Colleges
- Degree Programs
- Search Academics
PhD in Health Services & Policy Research
For contact information, please visit the School of Public Health website .
The Doctor of Philosophy in Health Services & Policy Research (PhD) degree program offered by the Department of Health Law, Policy & Management is designed to provide individuals with excellent research skills for use in academic, industry, or government settings. Students have the opportunity to collaborate with senior faculty in innovative research crucial to the improvement of healthcare delivery, treatment outcomes, and government policies. Graduates are known for their excellent methodological skills and substantive knowledge of healthcare settings and policies, competencies that enable students to translate research findings into practical applications.
The doctoral program supports a focus on quantitative methods and qualitative methods, while fostering facility with mixed methods designs.
Learning Outcomes
Upon completion of the PhD in Health Services & Policy Research (HSPR), the graduate is able to:
- Analyze key factors that have the potential to influence health and provision and use of health services. These may include policy, organization, and financing of healthcare services. They may also include social, economic, cultural, political, and biological factors that influence population health, health equity, and the use of services.
- Critique, adapt, and develop theoretical analyses of health services and policies, including explanations of their structures and processes, the use of health services, the effectiveness and implementation of health policies, and the effects of health services and policies on individual and population health and health equity. This may be done by drawing upon the foundational fields of sociology, economics, anthropology, political science, psychology, demography, epidemiology, management, and/or organizational science.
- Develop original, relevant, and important research questions to pursue in HSPR that are grounded in both a critical analysis of prior HSPR literature and relevant theoretical perspectives.
- Identify and analyze the strengths and weaknesses of a variety of possible study designs that can appropriately address specific HSPR questions. Appropriate methods may employ experimental or observational approaches, qualitative or quantitative analytic techniques, and a variety of data types.
- Based on relevant theory and selected research question(s), develop an HSPR design, specifying study constructs, research objectives, hypotheses, and methods that reliably and validly measure outcomes of interest. Select optimal analytical approaches, in combination as necessary, to investigate hypotheses.
- Acquire and manage appropriate data to answer HSPR questions. Collect and manage primary health and healthcare utilization data and/or assemble and manage existing data from public and private data sources in accordance with an original research design.
- Apply appropriate analytical techniques to data in order to investigate HSPR questions. Utilize appropriate combinations of analytic techniques to enrich data analysis and interpretation.
- Develop, document, and employ procedures that ensure the reproducibility of the science, the responsible use of resources, mutual accountability with collaborators, and the ethical treatment of research subjects.
- Effectively communicate the process, findings, and implications of HSPR using multiple modes, including via peer-reviewed publications, oral presentations, and electronic media. Translate findings to multiple stakeholders and audiences including funders, research participants, colleagues, policymakers, news media, and managers.
Program Requirements
The doctoral program includes the coursework requirements outlined below, which typically total at least 42 units. Students with an earned master’s degree may take fewer courses, if course waivers are granted based on previous relevant graduate coursework. However, in all cases, students are required to take no fewer than 32 units of PhD coursework in residence at Boston University post-matriculation into the PhD program. A minimum grade of a B is required in all PhD coursework.
Foundations of Public Health
Effective public health requires expertise from many disciplines, and students need to have a broad foundation of knowledge across these diverse disciplines in order to collaborate effectively with other health professionals.
SPH PH 700 Foundations of Public Health (0 units) is an online course designed to provide students with foundational knowledge in the profession and science of public health and factors related to public health. PH 700 meets the foundational knowledge criteria (as outlined by CEPH) for all SPH students.
Core PhD Courses
- SPH PH 842 Research Theory and Design (2 units)
- SPH PH 843 Quantitative Methods for Health and Social Policy Research (2 units)
- SPH PH 844 Introduction to Qualitative Methods for Health and Social Policy Research (2 units)
- SPH PM 820 Introduction to Quality Measurement and Evaluation (2 units)
- SPH PM 822 Advanced Quantitative Methods for Health and Social Policy Research (4 units)
- SPH PM 828 Advanced Qualitative Methods (4 units)
- SPH PM 842 Health Economics for Health and Social Policy Research or equivalent (4 units)
- SPH PM 846 Advanced Quantitative Policy and Program Implementation and Evaluation (4 units)
- SPH PM 864 Contemporary Structures of Health Services (2 units)
- SPH PM 866 Theory in the Analysis of Health Services (2 units)
Additional Course Requirements
- Healthcare Policy
- Implementation Science/Organizational Change
- Quality/Outcomes
Students are required to complete all the requirements for graduation within seven years of matriculation. PhD students are required to successfully complete the qualifying process, and complete and orally defend a dissertation.
Students who have completed the PhD coursework must register for SPH PM 980 Continuing Study each fall and spring term until the dissertation is defended and accepted. PM 980 is ungraded and 0 units; it allows a student to be certified as full time and carries the equivalent of a 2-unit tuition charge plus the fee for student health insurance (unless waived). Students must adhere to dissertation submission deadlines and requirements.
All PhD students must adhere to the Doctoral Graduation Calendar in preparing and submitting the dissertation.
All SPH students will need a laptop or tablet for classes, purchased according to the recommendations of the Medical Campus Information Technology professionals.
Related Bulletin Pages
- Abbreviations and Symbols
Beyond the Bulletin
- School of Public Health
- SPH Admissions
- SPH Academic Calendar
- Master of Public Health
- Health Equity
- Dual Degree Programs
- Business Administration & Public Health
- Genetic Counseling & Public Health MS/MPH
- Medical Sciences & Public Health MS/MPH
- Medicine & Public Health MD/MPH
- Social Work & Public Health MSW/MPH
- Doctor of Public Health in Leadership, Management & Policy
- Applied Biostatistics
- Biostatistics
- Climate and Health
- Environmental Health
- Epidemiology
- Global Health Research
- Health Services & Policy Research
- Population Health Research
- Public Health Data Science
- Translation and Implementation Science
- Boston Consortium
- Departments
Terms of Use
Note that this information may change at any time. Read the full terms of use .
related websites
Accreditation.
Boston University is accredited by the New England Commission of Higher Education (NECHE).

- © Copyright
- Mobile Version
- Degrees Offered
PhD in Health Services
Description.
The PhD Program in Health Services trains health services researchers and health policy analysts for careers in academic institutions, health delivery systems, public health departments, government agencies, and the private sector. The program prepares students to conduct high-quality independent, collaborative research and policy analysis by offering multidisciplinary, applied research opportunities on a wide variety of topics under the close mentorship of faculty. Students obtain advanced knowledge of the determinants of population health and of the health care system and they are exposed to several competing theoretical frameworks for conceptualizing both population health and the provision of health care. They develop research skills to identify and critically analyze the social, behavioral and health system effects on health and how organization, delivery, financing, and management of health services affect system performance.
Likely Careers
Health services researchers in academic institutions, health delivery systems, public health departments, government agencies, and the private sector, including insurance, pharmacy and biotechnology industries.
Applicants who have a bachelor, master, or professional degree in a field related to health services are given preference over applicants who do not have such experience.
Application Deadline: December 10, 2024
Application Deadline: Dec. 15, for Autumn Quarter entry
Competencies
Upon satisfactory completion of the PhD in Health Services, graduates will be able to:
- Meet the learning objectives of the PhD program in Health Services ;
- Demonstrate comprehensive understanding of the determinants, trends and major issues confronting U.S. health care/policy and its effect on individual and population health and health inequalities;
- Critically appraise journal articles, evaluate the evidence, synthesize findings, draw inferences and apply alternative theoretical and conceptual models from a range of relevant disciplines to health services (HS);
- Use knowledge of the structures, process/quality, performance, policy, and environmental context to formulate solutions for health and health care problems;
- Explain how to collect primary health and health care data obtained by survey, abstracts of medical records (including electronic medical records), and qualitative or mixed methods;
- Assemble secondary data from existing public and private sources and apply various statistical techniques to answer HS questions;
- Develop in-depth substantive/disciplinary knowledge and method skills in an area of emphasis and apply to health services problems demonstrating application of AoE to dissertation work;
- Ensure the ethical and responsible conduct of research in the design, implementation and dissemination of HS research;
- Critically appraise grants, understand the grant writing and review process, write research proposals, and Final Dissertation Proposal approved by faculty as part of the general examination;
- Work collaboratively in multidisciplinary teams;
- Conduct an independent HS research study of publishable quality, characterized by conceptual and methodological rigor, as well as practical value;
- Effectively communicate issues, research findings, and implications of HS through multiple modalities to appropriate professional, scientific, policy, and lay audiences;
- Collaborate with policy makers, organizations, and communities to plan, conduct and translate research into health policy and practice; and
- Participate and assist in teaching or clinical practice while in the program.

PhD in Health Services Research and Health Policy
Doctoral program in health services research and health policy, rollins school of public health department of health policy and management.
The PhD in Health Services Research and Health Policy at the Rollins School of Public Health at Emory University is a full-time program that trains researchers in the fields of health policy, health economics, health management, and health services research.
Students take doctoral-level classes in the Department of Economics, the Department of Health Policy and Management, the Goizueta business school, and elsewhere throughout the university. Many students also collaborate with faculty on research.
Following the completion of their coursework, students work on their independent research for their dissertation.
What You’ll Learn
Students in our program take classes in one of two tracks: Economics or Organizations and Management .
Economics Track
Students in the Economics track take graduate-level classes in the Department of Economics, alongside students pursing a PhD in economics. The economics track prepares students to apply economic theory to evaluate topics in health and health policy.
Organizations and Management Track
Students in Organizations and Management take advanced and doctoral-level courses in Emory’s Goizueta School of Business. The track prepares students to examine questions pertaining to access, quality, cost of health care and health outcomes. Students in this track will learn how theories and concepts from fields such as organizational behavior and technology management can be applied to medicine and health care organizations.
Core Courses and More
All students in the program take classes in statistical methods, research design, and health policy seminar. Students have room to take electives, which could be any graduate-level class at Emory or nearby universities (Georgia State, Georgia Tech).
For more information, please see our program brochure and handbook .
What Can You Do With a Graduate Degree in Health Services Research and Health Policy?
The program prepares students for a variety of research-focused careers in academia, think tanks, foundations, government agencies, pharmaceutical firms, and consulting.
Our graduates are currently employed at:
- the American Cancer Society
- the Centers for Disease Control and Prevention
- the U.S. Department of Health and Human Services
- Emory University
- Weill Cornell Medical School
- Harvard Medical School
- IMPAQ International
- Johnson & Johnson
- MD Anderson
- National Taiwan University
- Northern Illinois University
- Northwestern University
- Taipei Medical University
- The Urban Institute
- Trilliant Health
- University of California at San Francisco
- Carnegie Mellon
- Washington University (St. Louis)
- The University of Virginia
We discourage applications from students who view a PhD as a credential or who want to focus exclusively on administration, management, or advocacy. There are other professional degrees that are better suited to those types of careers.
What Type of Research Will You Do in the Health Services Research and Health Policy PhD Program?
Students perform research on a wide variety of topics related to delivery of medical care, insurance, and the determinants of health. Some examples of the papers that students have published from their dissertations include:
The effect of Medicaid expansion on crime reduction: Evidence from HIFA-waiver expansions. Journal of Public Economics 2017. (Heifei Wen, Ph.D. 2015)
Heuristics in the delivery room. Science 2021. (Manasvini Singh, Ph.D. 2020)
Are two heads better than one or do too many cooks spoil the broth? The tradeoff between physician division of labor and patient continuity of care for older adults with complex chronic conditions. Health Services Research 2016. (Kenton Johnston, Ph.D. 2015).
Effect of Medicaid disenrollment on health care utilization among adults with mental health disorders. Medical Care 2019 (Xu Ji, Ph.D. 2017).
Patterns of use and survival outcomes of positron emission tomography for initial staging in elderly follicular lymphoma patients . Leukemia & Lymphoma 2017 (Ashish Rai, Ph.D. 2015)
Admissions Requirements
For detailed information about admissions, please see our program brochure .
Applicants should provide:
- a transcript,
- statement of purpose,
- resume/CV, and
- three letters of recommendation.
Please note:
- GRE scores are optional.
- Applicants do not need to have a master’s degree.
Visit Emory’s Laney Graduate School website to apply now .
You do not need to contact the program or faculty prior to applying. We give equal attention to all applications, regardless of whether applicants know faculty or have had prior contact with them. We do not routinely meet with applicants prior to the application deadline. However, if you have a specific question about the program that is not addressed in this document or would like to get a better sense if the program is a good fit for you, please send your question to the director, David Howard, [email protected] .
September 11, 2023
Application opens for Fall 2024
December 1, 2023
Application deadline
Late January-Early February, 2024
Offer letters sent to successful applicants

Program faculty
Students have wide leeway to work with faculty at any Emory school or department. Most students work with the faculty on the list below.
Department of Health Policy and Management
Kathleen Adams (Ph.D. Economics, University of Colorado) Risk behavior, maternal and child health, insurance coverage, Medicaid policy.
Sarah Blake (Ph.D. Public Policy, Georgia State/Georgia Institute of Technology) Maternal and child health, reproductive health, implementation science.
Puneet Chehal (Ph.D. Public Policy, Duke) Medicaid and chronic illness in underserved populations.
Janet Cummings (Ph.D. Health Policy, UCLA) Mental health and substance abuse policy.
Benjamin Druss (M.D., New York University) Mental health and substance abuse policy.
Maria Dieci (Ph.D. Health Policy, UC Berkeley) Health economics, global health and development economics.
Ilana Graetz (Ph.D. Health Policy, UC Berkeley) Health information technology, quality improvement.
David Howard (Ph.D. Health Policy, Harvard) Health economics, reimbursement policy, pharmaceutical markets.
Joseph Lipscomb (Ph.D. Economics, University of North Carolina) Health outcomes assessment and improvement.
Victoria Phillips (Ph.D. Economics, Oxford) Health economics, cost-effectiveness analysis.
Adam Wilk (Ph.D. Health Policy, University of Michigan) Access to care and Medicaid, coverage and payment for kidney failure treatment.
Courtney Yarborough (Ph.D. Public Policy, University of Georgia) Substance abuse policy, pharmaceutical markets.
Affiliated faculty in other departments at Emory
Michal Hórny (Ph.D. Health Services Research, Boston University) Department of Radiology. Health insurance benefit design, costs of care, price transparency, access to care.
Xu Ji (Ph.D. Health Policy, Emory) Department of Pediatrics. Health care quality, health outcomes, access to health care.
Dio Kavalieratos (Ph.D. Health Policy, University of North Carolina) Department of Family Medicine and Palliative Care. End-of-life care, implementation science.
Sara Markowitz (Ph.D. Economics, CUNY) Department of Economics. Health economics, labor economics, maternal and child health. Ian McCarthy (Ph.D. Economics, University of Indiana) Department of Economics. Health economics, industrial organization.
Evan Saltzman (Ph.D. Managerial Science and Applied Economics, University of Pennsylvania) Department of Economics. Health economics, industrial organization.
Current PhD Students
Lamont Sutton
Nada Boualam
Sonia Tetlow
Xinyue Zhang
Marissa Coloske
Martha Wetzel
Paul George
Jingxuan Zhao
Alex Soltoff
Elizabeth Staton
Zhuoqi Yang
Cristian Ramos
Health Services Research and Policy

Our application for fall 2024 is now closed.
Health Services Research and Policy PhD Program
Since 1994, the PhD program in Health Services Research and Policy in the Department of Public Health Sciences has been highly successful in preparing students to become scholars for research, teaching, and public service careers in university, public policy, and governmental settings.
Our PhD program has also been designated as a STEM (Science, Technology, Engineering and Mathematics) program since 2010. Set within a large medical center with all its clinical research resources and collaborative faculty, our program focuses on interdisciplinary application of the social and behavioral science disciplines to real world health issues such as the organization, financing, and delivery of health care; the quality and safety of care; health outcomes; and the management of population health.
Read A Welcome Message from the Director
Program Highlights
- Our PhD program is designed to produce researchers who generate knowledge and strategies used in solving healthcare problems.
- Interdisciplinary courses, research and teaching, provide candidates with a broad range of skills.
- Our Ph.D. can be combined with other degrees such as the M.P.H. or M.D.
- Stipends, tuition grants, training and travel expenses are provided for doctoral study.
- The program prepares students for a career in academia, government or the private sector.
- Our PhD program exists within the Department of Public Health Sciences, which also houses MS programs in Epidemiology, Public Health, Health Services Research, and Clinical Investigations, and a PhD program in Epidemiology.
Current PhD Students
PhD Graduates
Primary Faculty
Publications per year
What Sets Us Apart

Information on core courses, electives, lab rotations to guide you through our program

We support and guide trainees in several areas of research. Browse research by current program trainees

Mentor Relationships
Our program faculty offer a wide range of research experience to mentor you towards your Ph.D.
Discover Public Health Sciences
Latest News
August 8, 2024 Dr. Jose Perez Ramos Co-Authors AJPH Supplement on Latino Health Inequities
August 8, 2024 Misti Ellinger and Rochester Coalition Highlight Breastfeeding’s Public Health Impact
August 1, 2024 Dr. Scott McIntosh was interviewed for recent news article
Upcoming Events
There is no upcoming seminar scheduled.
All Events »
What Our Students Say...

"I chose the University of Rochester's Health Services Research and Policy program because the faculty and staff provide students with a supportive environment to develop their research ideas. The program also has a flexible curriculum. Students can take classes offered by other departments- i.e., data science, psychology."
Hometown: Zhejiang, China
Undergraduate Degree: Finance
Areas of Interest: Health Economics; Price Transparency in Healthcare; Quality and Cost; Econometric Modeling
For information related to the doctoral degree program in Health Services Research and Policy, please contact:
Yue Li, PhD , Director
Health Services Research and Policy Doctoral Program Department of Public Health Sciences University of Rochester Medical Center
Office: (585) 275-3276 Fax: (585) 461-4532
Connect With Us On Social Media
- Skip to main content
- Skip to main navigation
PhD in Health Services Research
Become a thought leader in the healthcare profession.
The purpose of the PhD program in Health Services Research in the Hankamer School of Business is to train researchers in cutting edge data analysis skills. The individuals completing the program will primarily work in large health systems and provide detailed and rigorous analysis that will help managers make decisions to improve the quality and/or lower the cost of health services delivered. Why a Baylor PhD?
Distinctives
The PhD Program is unique among many health services and health policy oriented doctoral programs as it is designed with the inclusion of a Master of Science (MS) in Economics degree to be completed prior to entering the dissertation phase of the program. This component of the PhD curriculum reflects the strong emphasis on economic theory and empirical techniques to guide students' research.

The Hankamer School of Business is also home to other health care-focused academic endeavors, including the Robbins Institute for Health Policy and Leadership , which supports the schools’ healthcare MBA program. Additionally, Baylor is a proud member of the Business School Alliance for Health Management ( BAHM ), a consortium of business-school healthcare programs at 17 leading universities.
BAHM journal: Health Management, Policy, and Innovation
Is This the Program for You?
Our program is highly selective, and most or all students receive considerable financial support. Admission with an offer of financial support typically requires a strong academic record — ideally with considerable training in Statistics, Mathematics, and Economics. If you have this background and a strong desire to acquire cutting-edge research skills and use them to improve health care delivery in the US and around the world, we encourage you to apply. Learn more about our admissions processes .
More About Our Faculty:
- Featured Publications
Program Directors
Dr. Michael Richards [email protected]
Dr. Scott Cunningham [email protected]
- Hankamer School of Business
Paul L. Foster Campus for Business and Innovation 1621 S 3rd St. Waco, TX 76706
One Bear Place #98001 Waco, TX 76798
- General Information
- Academics & Research
- Administration
- Gateways for ...
- About Baylor
- Give to Baylor
- Social Media
- Strategic Plan
- College of Arts & Sciences
- Diana R. Garland School of Social Work
- George W. Truett Theological Seminary
- Graduate School
- Honors College
- Louise Herrington School of Nursing
- Research at Baylor University
- Robbins College of Health and Human Sciences
- School of Education
- School of Engineering & Computer Science
- School of Music
- University Libraries, Museums, and the Press
- More Academics
- Compliance, Risk and Safety
- Human Resources
- Marketing and Communications
- Office of General Counsel
- Office of the President
- Office of the Provost
- Operations, Finance & Administration
- Senior Administration
- Student Life
- University Advancement
- Undergraduate Admissions
- Graduate Admissions
- Baylor Law School Admissions
- Social Work Graduate Programs
- George W. Truett Theological Seminary Admissions
- Online Graduate Professional Education
- Virtual Tour
- Visit Campus
- Alumni & Friends
- Faculty & Staff
- Prospective Faculty & Staff
- Prospective Students
- Anonymous Reporting
- Annual Fire Safety and Security Notice
- Cost of Attendance
- Digital Privacy
- Legal Disclosures
- Mental Health Resources
- Web Accessibility
- Request Info
- Departments
- Community Service
- Patient Care
- Give to SLU Medicine
- Search & Directory
Health Outcomes Research, Ph.D. and Medicine, M.D. Dual Degree
- Requirements
- Contact Info
Saint Louis University's M.D./Ph.D. program in Health Outcomes Research stands at the forefront of training physician-scientists to tackle complex healthcare issues. This innovative dual-degree program integrates intensive medical education with advanced research training, empowering students to investigate and address critical questions in healthcare delivery, patient outcomes, and healthcare policy.
Students in this program benefit from a comprehensive curriculum that combines clinical rotations with rigorous research methodologies. They engage in cutting-edge research projects under the mentorship of esteemed faculty, gaining hands-on experience in analyzing health data, conducting clinical trials, and developing innovative approaches to improve healthcare quality and accessibility.
Graduates of SLU's M.D./Ph.D. program emerge with a deep understanding of both clinical practice and scientific inquiry, equipped to drive impactful change in healthcare systems. They are prepared to lead interdisciplinary teams, collaborate across specialties, and translate research findings into tangible improvements in patient care and healthcare policy.
With access to state-of-the-art facilities, a supportive academic community, and opportunities to collaborate with leading healthcare organizations, SLU's M.D./Ph.D. program in Health Outcomes Research empowers future physician-scientists to make meaningful contributions to the field, shaping the future of healthcare for generations to come.
For additional information, see the catalog entries for the following SLU programs:
Medicine, M.D.
Health Outcomes Research, Ph.D.
Scholarships and Financial Aid
For priority consideration for graduate assistantship, apply by Feb. 1.
For more information, visit the Office of Student Financial Services .
Accreditation
Saint Louis University is accredited by the Higher Learning Commission (HLC) and has been continuously accredited since 1916.
Admission Requirements
Students must first be admitted to the School of Medicine before being considered for acceptance into the M.D./Ph.D. in Health Outcomes Research. Questions about admission to the dual-degree program can be directed to the M.D./Ph.D. program director.
Most trainees are chosen in the early spring to begin their research training by mid-June before commencing their first year of M.D. studies in mid-August. Other qualified individuals may apply after starting their M.D. training.
Applicants to the M.D./Ph.D. program are screened by the steering committee for the M.D./Ph.D. program.
Program Requirements
Students begin their program by completing the first and second years of their M.D. studies. By the winter of their second year of medical studies, students will select a Ph.D. mentor from SLU’s Department of Health and Clinical Outcomes Research.
Completion of all Ph.D. candidacy requirements and oral defense of the doctoral dissertation is followed by a transitional clerkship that facilitates re-entry into the third and fourth years of M.D. studies.
Up to 9 credits, with approval from Ph.D. mentor, from the M.D. program will count towards the completion of the Ph.D. in Health Outcomes Research.
Trainees in this program should complete all requirements for both degrees within seven to eight years and students are expected to complete both the M.D. and Ph.D. components on a full-time basis.
Continuation Standards
Students must maintain a cumulative grade point average (GPA) of 3.00 in all graduate/professional courses.
For more information about the health outcomes research Ph.D. or this degree program, contact:
Department of Health and Clinical Outcomes Research [email protected]
- Request Info
- Departments
- Community Service
- Patient Care
- Give to SLU Medicine
- Search & Directory
Health Outcomes Research, Ph.D.
- Learning Outcomes
- Requirements
Health outcomes research is a rapidly expanding, interdisciplinary field that provides evidence and guidance for understanding the endpoints of treatments, interventions and health care practices, be they clinical, functional, quality-of-life or economic. Saint Louis University’s Doctor of Philosophy (Ph.D.) in Health Outcomes Research is a program that trains researchers in the areas of health outcomes research, health services research and health data science to meet the changing needs of the health care system.
The goal of SLU's program is to prepare robust clinical and health outcomes researchers. Students receive a solid foundation in:
- Research methodology
- Data management
- Statistical analysis
- Data science
- Scientific writing and presentation
In addition to coursework, students work collaboratively with their mentor, research and clinical faculty and other students to produce high-quality research throughout their program.
Curriculum Overview
The program requires a total of 48 credits for completion; 36 credits of coursework and 12 dissertation credits. Additionally, students must pass a written comprehensive exam, an oral examination/proposal of the dissertation and a public presentation and defense of the dissertation.
Fieldwork and Research Opportunities
The department partners with clinical faculty in the SLU School of Medicine and conducts research in numerous clinical areas, including diabetes, oncology, pediatrics, otolaryngology, infectious disease and health care quality. Additionally, our faculty have expertise in health data science, research methodology, biostatistics, epidemiology, survey design and outcomes measurement. Students have the opportunity to work with their primary mentor, our faculty and clinical faculty on both short- and long-term research projects.
Graduates are prepared to work as academics and researchers at universities, medical centers, government and nongovernment health agencies, hospital systems, insurance and other areas of the health industry.
Admission Requirements
Applicants should have a master’s degree from an accredited college or university in social science, biomedical science, public health, or related discipline. Successful candidates will have maintained a minimum 3.5 GPA in graduate coursework and scored at least at the 50th percentile for GRE verbal and quantitative reasoning. Students must also demonstrate evidence of interest in an area of research and identify a willing and suitable faculty mentor.
Application Requirements
Begin your application for this program at www.slu.edu/apply .
- Application form and fee
- Transcripts from most recent degree(s)
- Professional statement
- Résumé or curriculum vitae
- One letter of recommendation
- GRE required
Requirements for International Students
Along with the general admission requirements above, the following must be provided by prospective international students:
- Demonstration of English Language Proficiency .
- A letter of financial support from the person(s) or sponsoring agency funding the time at Saint Louis University.
- A letter from the sponsor's bank verifying that the funds are available and will be so for the duration of study at the University.
- Academic records, in English translation, for postsecondary studies outside the United States. These must include the courses taken and/or lectures attended, practical laboratory work, the maximum and minimum grades attainable, the grades earned or the results of all end-of-term examinations, and any honors or degrees received. WES and ECE transcripts are accepted.
Application Deadline
Applications to the program are considered on a rolling basis.
| Tuition | Cost Per Credit |
|---|---|
| Graduate Tuition | $1,370 |
Additional charges may apply. Other resources are listed below:
Net Price Calculator
Information on Tuition and Fees
Miscellaneous Fees
Information on Summer Tuition
Scholarships and Financial Aid
For priority consideration for graduate assistantship, apply by Feb. 1.
For more information, visit the Office of Student Financial Services .
Accreditation
Saint Louis University is accredited by the Higher Learning Commission (HLC) and has been continuously accredited since 1916.
- Graduates will be able to effectively review, summarize and synthesize literature related to clinical aspects of health outcomes.
- Graduates will be able to apply appropriate data management strategies related to clinical aspects of health outcomes.
- Graduates will be able to critically evaluate clinical aspects and health care-specific methodological designs.
- Graduates will be able to demonstrate a thorough and ethical approach to conducting academic research.
- Graduates will be able to effectively communicate study results related to clinical aspects of health outcomes.
| Code | Title | Credits |
|---|---|---|
| Required Courses | ||
| ORES 5010 | Introduction to Biostatistics for Health Outcomes | 3 |
| or HDS 5310 | Analytics and Statistical Programming | |
| ORES 5160 | Data Management | 3 |
| ORES 5300 | Foundations of Outcomes Research I | 3 |
| ORES 5320 | Scientific Writing and Communication | 3 |
| ORES 5430 | Health Outcomes Measurement | 3 |
| ORES 5150 | 0-3 | |
| or HDS 5320 | Inferential Modeling | |
| ORES 6990 | Dissertation Research (taken over multiple semesters, 12hrs total) | 0-6 |
| Program Elective Courses | ||
| Select six courses from the following: | 18 | |
| Programming for Health Data Scientists | ||
| High Performance Computing | ||
| Predictive Modeling and Machine Learning | ||
| Health Care Organization | ||
| Foundations of Medical Diagnosis and Treatment | ||
| Pharmacoeconomics | ||
| Evaluation Sciences | ||
| R Programming | ||
| SAS Programming I | ||
| Pharmacoepidemiology | ||
| Comparative Effectiveness Research | ||
| Advanced Graduate Readings in Outcomes Research | ||
| Total Credits | 45-48 | |
Continuation Standards
Students must maintain a cumulative grade point average (GPA) of 3.00 in all graduate/professional courses.
Roadmaps are recommended semester-by-semester plans of study for programs and assume full-time enrollment unless otherwise noted.
Courses and milestones designated as critical (marked with !) must be completed in the semester listed to ensure a timely graduation. Transfer credit may change the roadmap.
This roadmap should not be used in the place of regular academic advising appointments. All students are encouraged to meet with their advisor/mentor each semester. Requirements, course availability and sequencing are subject to change.
| Year One | ||
|---|---|---|
| Fall | Credits | |
| ORES 5010 | Introduction to Biostatistics for Health Outcomes | 3 |
| ORES 5300 | Foundations of Outcomes Research I | 3 |
| ORES 5320 | Scientific Writing and Communication | 3 |
| Credits | 9 | |
| Spring | ||
| ORES 5160 | Data Management | 3 |
| ORES 5210 | Foundations of Medical Diagnosis and Treatment (Program Elective #1) | 3 |
| HDS 5210 | Programming for Health Data Scientists (Program Elective #2) | 3 |
| Credits | 9 | |
| Summer | ||
| HDS 5320 | Inferential Modeling (Can substitute for ORES 5150 or be used as an elective) | 3 |
| Credits | 3 | |
| Year Two | ||
| Fall | ||
| ORES 5430 | Health Outcomes Measurement | 3 |
| Program Elective #3 | 3 | |
| Program Elective #4 | 3 | |
| Credits | 9 | |
| Spring | ||
| Program Elective #5 | 3 | |
| Program Elective #6 | 3 | |
| Credits | 6 | |
| Year Three | ||
| Fall | ||
| ORES 6990 | Dissertation Research | 6 |
| Credits | 6 | |
| Spring | ||
| ORES 6990 | Dissertation Research | 6 |
| Credits | 6 | |
| Total Credits | 48 | |


K. Davina Frick, PhD
| Academic Area | Economics |
|---|---|
| Academic Area | Health |
| Areas of Interest | Economics, Healthcare Management, Leadership and Values |
|---|
K. Davina Frick (she/her) is a professor who teaches economics for decision-making, business leadership and human values, frameworks for analyzing healthcare markets, and a course on how the U.S. health care system in the past, present, and future facilitates innovation.
Frick studied health policy and administration at Penn State, followed by economics and health services research at the University of Michigan. In 1996, she joined the faculty of the Johns Hopkins Bloomberg School of Public Health, where she still has joint appointments. She moved to a leadership position at the Carey Business School in 2013 and returned to an exclusively faculty role in 2021.
Much of Frick’s research focuses on measuring costs associated with diseases or measuring the cost-effectiveness of new treatments, care systems, or community-based interventions. The most focused area has been public and private eye care, and she is currently co-chairing a National Academies of Science, Engineering, and Medicine workgroup on myopia. In addition, she is co-chairing an AcademyHealth project on health services research’s inclusivity, impact, and innovation. She focuses on research translation, specializing in linking peer-reviewed research with the information business leaders need to make decisions about companies that operate in the real-world marketplace. She plans for future research to focus on mentoring, leadership, and DEIB issues.
Frick has mentored students, faculty, and staff, and spoken about mentoring to many audiences—particularly mentoring as a two-way street. She has focused on DEIB issues as a member of a team that produced a series of videos about LGBTQ+ Narratives in Academia, as part of a second team that produced a video called Business of Pronouns, by serving on the school’s Inclusive Teaching committee as well as the university’s Diversity Leadership Council, by reading names at graduation for 10 years, and by serving on the Diversity and Anti-Racism Team for the Society for Medical Decision Making.
- Ph. D, Economics and Health Services Organization and Policy, University of Michigan
- MA, Economics, University of Michigan
- BS, Health Policy & Administration, Pennsylvania State University
Selected publications
- Garcia J, Yesantharao L, Frick KD, Fakhry C, Koch W, Mydlarz W, Eisele DW, Gourin CG. Concentration of high-cost head and neck cancer surgical patients. d and neck cancer surgical patients. The Laryngoscope . Forthcoming.
- Markoulli M, Fricke T R, Arvind A, Frick KD, Hart KM, Joshi M., Kandel H, Filipe Macedo A, Makrynioti D, Retallic N, Garcia-Porta N, Shrestha G, Wolffsohn JS. (2024). BCLA CLEAR Presbyopia: Epidemiology and impact. Contact lens & anterior eye : the journal of the British Contact Lens Association , 102157. Advance online publication. https://doi.org/10.1016/j.clae.2024.102157
- Wong B, Singh K, Everett B, O’Brien KS, Ravilla T, Khanna RC, Chase H, Frick KD. The Potential of Eye Health Investment as a Best Buy in Global Health and Development: A Systematic Review and Economic Modeling Analysis. Bulletin of the World Health Organization . Forthcoming.
- Sun J, Frick KD, Liang H, Chow CM, Aronwitz S, Shi L. Examining cancer screening disparities by race/ethnicity and insurance groups: A comparison of 2008 and 2018 National Health Interview Survey (NHIS) data in the United States. PLOS ONE . Forthcoming.
- Saraswathula A, Yesantharao ., Gourin CG, Rowan NR, Frick KD. (2023). Cost-effectiveness analysis comparing in-office posterior nasal nerve ablation to surgical therapies. American Journal of Otolaryngology , 44(2), 103776. https://doi.org/10.1016/j.amjoto.2022.103776.
- Wong B, Singh K, Khanna RC, Ravilla T, Kuyyadiyil S, Sabherwal S, Sil A, Dole K, Chase H, Frick KD. Strategies for cataract and uncorrected refractive error case finding in India: costs and cost-effectiveness at scale. The Lancet Regional Health—Southeast Asia. Forthcoming.
- Miller, J. R., Frick, K. D., & Gourin, C. G. (2022). Hospital Markup in Head and Neck Cancer Surgery in the US. JAMA otolaryngology-- head & neck surgery. 148(12): 1147–1155. https://doi.org/10.1001/jamaoto.2022.3340.
- Saraswathula, A., Austin, J. M., Fakhry, C., Vosler, P. S., Mandal, R., Koch, W. M., Tan, M., Eisele, D. W., Frick, K. D., & Gourin, C. G. (2023). Surgeon Volume and Laryngectomy Outcomes . The Laryngoscope , 133(4), 834–840.
Working Papers
- Segal J, Yanek L, Jager L, Okoli E, Hatef E, Dada M, Frick KD. Higher percentage of virtual primary care associated with differences in achievement of some quality metrics.
- Wong B, Singh K, Ravilla T, Khanna RC, Chase H, Frick KD.The Potential of Eye Health Investment as a Best Buy in Global Health and Development.
- Collins ME, Alexander G, Guo X, Tariq A, Frick KD. Cost Analysis of a School-Based Vision Program in an Urban, High-Poverty School District.
- Frameworks for Analyzing Health Care Markets
- Business Leadership and Human Values
- U.S. Health Care System: Past, Present, and Future
- Economics for Decision Making
Honors and distinctions
- Penn State Alumni Association, Martin R. Cepeda, Jr. Award for Alumni Career Advancement and Development, (August 2024)
- Johns Hopkins University Diversity Recognition Award (2024) Johns Hopkins Carey Business School (2023)
- Collaborative Leadership Award Johns Hopkins University Diversity Recognition Award (2022)
- Interviewer and interview annotator for the LGBTQ+ Narratives in Academia Project Journal of Nutrition Education and Behavior, Best Article Award (2017).
- Co-author on “APHA Vision Care Section Distinguished Service (2011)
- Golden Apple Award for Small Class Sizes (2011), Johns Hopkins Bloomberg School of Public Health Student Assembly
- Golden Apple Award for Internet-based Classes (2011), Johns Hopkins Bloomberg School of Public Health
- Student Assembly Penn State Schreyer Honors College (2009) Outstanding Scholar Alumnus
Impacts and Engagements
- Editor, Sage Knowledge Healthcare Management Series (2024-Present)
- Associate Editor, Women’s Health Issues (2020-Present)
- Sponsored and participated in a panel discussion following the presentation of the documentary You Belong Here (about coming out) at the Johns Hopkins University School of Nursing
- Spoke about developing a joy-centered personal mission statement at the National Conference for College Women Student Leaders
- Spoke about health care economics at an NIDCD/FDA Working Group on Accessible and Affordable Hearing Health Care for Adults
Multiple podcasts and blogs on leadership, values, and DEIB
- DEIB Summit: ERGs & Affinity-Based...
- HCA Healthcare Info Session
- Industry Insights & Connections with Alumni in...
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Review Article
- Published: 22 March 2024
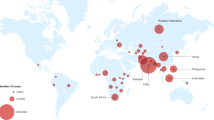
Drug-resistant tuberculosis: a persistent global health concern
- Maha Farhat ORCID: orcid.org/0000-0002-3871-5760 1 , 2 ,
- Helen Cox 3 na1 ,
- Marwan Ghanem 1 na1 ,
- Claudia M. Denkinger 4 , 5 ,
- Camilla Rodrigues 6 ,
- Mirna S. Abd El Aziz 4 ,
- Handaa Enkh-Amgalan 7 ,
- Debrah Vambe 8 ,
- Cesar Ugarte-Gil 9 ,
- Jennifer Furin 10 &
- Madhukar Pai ORCID: orcid.org/0000-0003-3667-4536 11
Nature Reviews Microbiology ( 2024 ) Cite this article
6265 Accesses
3 Citations
165 Altmetric
Metrics details
- Antimicrobial resistance
- Bacterial evolution
- Clinical microbiology
- Infectious-disease epidemiology
Drug-resistant tuberculosis (TB) is estimated to cause 13% of all antimicrobial resistance-attributable deaths worldwide and is driven by both ongoing resistance acquisition and person-to-person transmission. Poor outcomes are exacerbated by late diagnosis and inadequate access to effective treatment. Advances in rapid molecular testing have recently improved the diagnosis of TB and drug resistance. Next-generation sequencing of Mycobacterium tuberculosis has increased our understanding of genetic resistance mechanisms and can now detect mutations associated with resistance phenotypes. All-oral, shorter drug regimens that can achieve high cure rates of drug-resistant TB within 6–9 months are now available and recommended but have yet to be scaled to global clinical use. Promising regimens for the prevention of drug-resistant TB among high-risk contacts are supported by early clinical trial data but final results are pending. A person-centred approach is crucial in managing drug-resistant TB to reduce the risk of poor treatment outcomes, side effects, stigma and mental health burden associated with the diagnosis. In this Review, we describe current surveillance of drug-resistant TB and the causes, risk factors and determinants of drug resistance as well as the stigma and mental health considerations associated with it. We discuss recent advances in diagnostics and drug-susceptibility testing and outline the progress in developing better treatment and preventive therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
195,33 € per year
only 16,28 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Multidrug-resistant tuberculosis
Rifampicin Resistant Tuberculosis in Lesotho: Diagnosis, Treatment Initiation and Outcomes

Global burden of disease due to rifampicin-resistant tuberculosis: a mathematical modeling analysis
Peto, H. M., Pratt, R. H., Harrington, T. A., LoBue, P. A. & Armstrong, L. R. Epidemiology of extrapulmonary tuberculosis in the United States, 1993–2006. Clin. Infect. Dis. 49 , 1350–1357 (2009).
Article PubMed Google Scholar
WHO Consolidated Guidelines on Tuberculosis. Module 4: Treatment — Drug-resistant Tuberculosis Treatment, 2022 Update. World Health Organization https://www.who.int/publications/i/item/9789240063129 (2022).
WHO Consolidated Guidelines on Tuberculosis. Module 4: Treatment — Drug-susceptible Tuberculosis Treatment. World Health Organization https://www.who.int/publications/i/item/9789240048126 (2022).
WHO Consolidated Guidelines on Tuberculosis. Module 3: Diagnosis — Rapid Diagnostics For Tuberculosis Detection, 2021 Update. World Health Organization https://www.who.int/publications/i/item/9789240029415 (2021).
Cohen, K. A., Manson, A. L., Desjardins, C. A., Abeel, T. & Earl, A. M. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med. 11 , 45 (2019).
Article PubMed PubMed Central Google Scholar
Cohen, K. A. et al. Extensive global movement of multidrug-resistant Mycobacterium tuberculosis strains revealed by whole-genome analysis. Thorax 74 , 882–889 (2019).
Short-course chemotherapy in pulmonary tuberculosis: a controlled trial by the British Thoracic and Tuberculosis Association. Lancet 305 , 119–124 (1975).
Zhang, Y. The magic bullets and tuberculosis drug targets. Annu. Rev. Pharmacol. Toxicol. 45 , 529–564 (2005).
Article CAS PubMed Google Scholar
Global Tuberculosis Report 2022. World Health Organization https://www.who.int/publications/i/item/9789240061729 (2022).
Global Tuberculosis Report 2021. World Health Organization https://www.who.int/publications/i/item/9789240037021 (2021).
Boehme, C. C. et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363 , 1005–1015 (2010).
Article CAS PubMed PubMed Central Google Scholar
The Use of Molecular Line Probe Assay for the Detection of Resistance to Isoniazid and Rifampicin: Policy Update. World Health Organization https://www.who.int/publications/i/item/9789241511261 (2016).
Dixit, A. et al. Estimation of country-specific tuberculosis antibiograms using genomic data. Preprint at medRxiv https://doi.org/10.1101/2021.09.23.21263991 (2021).
Article Google Scholar
O’Connor, C. & Brady, M. F. Isoniazid. StatPearls [Internet] https://pubmed.ncbi.nlm.nih.gov/32491549/ (updated 8 April 2022).
Yee, D. et al. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am. J. Respir. Crit. Care Med. 167 , 1472–1477 (2003).
Murray, J. F., Schraufnagel, D. E. & Hopewell, P. C. Treatment of tuberculosis: a historical perspective. Ann. Am. Thorac. Soc. 12 , 1749–1759 (2015).
Jacobson, K. R. et al. Treatment outcomes of isoniazid-resistant tuberculosis patients, Western Cape Province, South Africa. Clin. Infect. Dis. 53 , 369–372 (2011).
Ahmad, N., Ahuja, S. & Akkerman, O. Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet 392 , 821–834 (2018).
Stagg, H. R. et al. Fluoroquinolones and isoniazid-resistant tuberculosis: implications for the 2018 WHO guidance. Eur. Respir. J. 54 , 1900982 (2019).
WHO Treatment Guidelines for Isoniazid-resistant Tuberculosis: Supplement to the WHO Treatment Guidelines for Drug-resistant Tuberculosis. World Health Organization https://www.who.int/publications/i/item/9789241550079 (2018).
Meeting Report of the WHO Expert Consultation on the Definition of extensively Drug-Resistant Tuberculosis, 27-29 October 2020. World Health Organization https://www.who.int/publications/i/item/9789240018662 (2021).
Global Tuberculosis Report 2023. World Health Organization https://www.who.int/publications/i/item/9789240083851 (2023).
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 , 629–655 (2022).
Article CAS Google Scholar
Knight, G. M., McQuaid, C. F., Dodd, P. J. & Houben, R. Global burden of latent multidrug-resistant tuberculosis: trends and estimates based on mathematical modelling. Lancet Infect. Dis. 19 , 903–912 (2019).
WHO Global Task Force on TB Impact Measurement: Report of a Subgroup Meeting on Methods Used by WHO to Estimate TB Disease Burden, 11-12 May 2022, Geneva, Switzerland. World Health Organization https://www.who.int/publications/i/item/9789240057647 (2022).
Global Tuberculosis Report 2015. World Health Organization https://www.who.int/publications/i/item/9789241565059 (2015).
Global Tuberculosis Report 2020. World Health Organization https://www.who.int/publications/i/item/9789240013131 (2020).
Villegas, L. et al. Prevalence, risk factors, and treatment outcomes of isoniazid- and rifampicin-mono-resistant pulmonary tuberculosis in Lima, Peru. PLoS ONE 11 , e0152933 (2016).
Sharling, L., Marks, S. M., Goodman, M., Chorba, T. & Mase, S. Rifampin-resistant tuberculosis in the United States, 1998-2014. Clin. Infect. Dis. 70 , 1596–1605 (2019).
Ismail, N. A. et al. Prevalence of drug-resistant tuberculosis and imputed burden in South Africa: a national and sub-national cross-sectional survey. Lancet Infect. Dis. 18 , 779–787 (2018).
Dean, A. S. Prevalence and genetic profiles of isoniazid resistance in tuberculosis patients: a multicountry analysis of cross-sectional data. PLoS Med. 17 , e1003008 (2020).
Subbaraman, R., Jhaveri, T. & Nathavitharana, R. R. Closing gaps in the tuberculosis care cascade: an action-oriented research agenda. J. Clin. Tuberc. Mycobact. Dis. 19 , 100144 (2020).
Google Scholar
Subbaraman, R. et al. Constructing care cascades for active tuberculosis: a strategy for program monitoring and identifying gaps in quality of care. PLoS Med. 16 , e1002754 (2019).
Naidoo, P. et al. The South African tuberculosis care cascade: estimated losses and methodological challenges. J. Infect. Dis. 216 , S702–S713 (2017).
Subbaraman, R. et al. The tuberculosis cascade of care in India’s public sector: a systematic review and meta-analysis. PLoS Med. 13 , e1002149 (2016).
Migliori, G. B. et al. Gauging the impact of the COVID-19 pandemic on tuberculosis services: a global study. Eur. Respir. J. 58 , 2101786 (2021).
Daniels, B. et al. Use of standardised patients to assess quality of healthcare in Nairobi, Kenya: a pilot, cross-sectional study with international comparisons. BMJ Glob. Health 2 , e000333 (2017).
Daniels, B., Kwan, A. & Pai, M. Lessons on the quality of tuberculosis diagnosis from standardized patients in China, India, Kenya, and South. Afr. J. Clin. Tuberc. Mycobact. Dis. 16 , 100109 (2019).
Boffa, J. et al. Quality of care for tuberculosis and HIV in the private health sector: a cross-sectional, standardised patient study in South Africa. BMJ Glob. Health 6 , e005250 (2021).
Kwan, A. et al. Variations in the quality of tuberculosis care in urban India: a cross-sectional, standardized patient study in two cities. PLoS Med. 15 , e1002653 (2018).
Daniels, B. et al. Tuberculosis diagnosis and management in the public versus private sector: a standardised patients study in Mumbai, India. BMJ Glob. Health 7 , 009657 (2022).
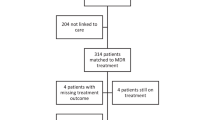
Demers, A. M. et al. Drug susceptibility patterns of Mycobacterium tuberculosis from adults with multidrug-resistant tuberculosis and implications for a household contact preventive therapy trial. BMC Infect. Dis. 21 , 205 (2021).
Step up for TB 2020 report: Tuberculosis Policies in 37 Countries. Médecins Sans Frontières & Stop TB Partnership https://msfaccess.org/step-tb-tb-policies-37-countries-4th-ed (2020).
Omar, S. V., Ismail, F., Ndjeka, N., Kaniga, K. & Ismail, N. A. Bedaquiline-resistant tuberculosis associated with Rv0678 mutations. N. Engl. J. Med. 386 , 93–94 (2022).
Ismail, N. A. et al. Assessment of epidemiological and genetic characteristics and clinical outcomes of resistance to bedaquiline in patients treated for rifampicin-resistant tuberculosis: a cross-sectional and longitudinal study. Lancet Infect. Dis. 22 , 496–506 (2022).
Azimi, T. et al. Linezolid resistance in multidrug-resistant Mycobacterium tuberculosis : a systematic review and meta-analysis. Front. Pharmacol. 13 , 955050 (2022).
Chesov, E. et al. Emergence of bedaquiline resistance in a high tuberculosis burden country. Eur. Respir. J. 59 , 2100621 (2022).
Mallick, J. S., Nair, P., Abbew, E. T., Van Deun, A. & Decroo, T. Acquired bedaquiline resistance during the treatment of drug-resistant tuberculosis: a systematic review. JAC Antimicrob. Resist. 4 , dlac029 (2022).
Jenkins, H. E. & Yuen, C. M. The burden of multidrug-resistant tuberculosis in children. Int. J. Tuberc. Lung Dis. 22 , 3–6 (2018).
WHO Consolidated Guidelines on Tuberculosis, Module 5: Management of Tuberculosis in children and Adolescents. World Health Organization https://www.who.int/publications/i/item/9789240046764 (2022).
Dodd, P. J., Mafirakureva, N., Seddon, J. A. & McQuaid, C. F. The global impact of household contact management for children on multidrug-resistant and rifampicin-resistant tuberculosis cases, deaths, and health-system costs in 2019: a modelling study. Lancet Glob. Health 10 , 1034–1044 (2022).
Dookie, N., Rambaran, S., Padayatchi, N., Mahomed, S. & Naidoo, K. Evolution of drug resistance in Mycobacterium tuberculosis : a review on the molecular determinants of resistance and implications for personalized care. J. Antimicrob. Chemother. 73 , 1138–1151 (2018).
Casali, N. et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46 , 279–286 (2014).
Cohen, K. A. et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 12 , e1001880 (2015).
Jiang, Q. et al. The evolution and transmission dynamics of multidrug-resistant tuberculosis in an isolated high-plateau population of Tibet, China. Microbiol. Spectr. 11 , e03991-22 (2023).
Ektefaie, Y., Dixit, A., Freschi, L. & Farhat, M. R. Globally diverse Mycobacterium tuberculosis resistance acquisition: a retrospective geographical and temporal analysis of whole genome sequences. Lancet Microbe 2 , e96–e104 (2021).
Auld, S. C. et al. Extensively drug-resistant tuberculosis in South Africa: genomic evidence supporting transmission in communities. Eur. Respir. J. 52 , 1800246 (2018).
Kendall, E. A., Fofana, M. O. & Dowdy, D. W. Burden of transmitted multidrug resistance in epidemics of tuberculosis: a transmission modelling analysis. Lancet Respir. Med. 3 , 963–972 (2015).
Yang, C. et al. Transmission of multidrug-resistant Mycobacterium tuberculosis in Shanghai, China: a retrospective observational study using whole-genome sequencing and epidemiological investigation. Lancet Infect. Dis. 17 , 275–284 (2017).
Becerra, M. C. et al. Transmissibility and potential for disease progression of drug resistant Mycobacterium tuberculosis : prospective cohort study. BMJ 367 , l5894 (2019).
Atre, S. R. et al. Tuberculosis pathways to care and transmission of multidrug resistance in India. Am. J. Respir. Crit. Care Med. 205 , 233–241 (2022).
El Halabi, J. et al. Measuring health-care delays among privately insured patients with tuberculosis in the USA: an observational cohort study. Lancet Infect. Dis. 21 , 1175–1183 (2021).
Odone, A. et al. Acquired and transmitted multidrug resistant tuberculosis: the role of social determinants. PLoS ONE 11 , e0146642 (2016).
Bayer, R. & Wilkinson, D. Directly observed therapy for tuberculosis: history of an idea. Lancet 345 , 1545–1548 (1995).
Pasipanodya, J. G., Srivastava, S. & Gumbo, T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of antituberculosis therapy. Clin. Infect. Dis. 55 , 169–177 (2012).
Srivastava, S., Pasipanodya, J. G., Meek, C., Leff, R. & Gumbo, T. Multidrug-resistant tuberculosis not due to noncompliance but to between-patient pharmacokinetic variability. J. Infect. Dis. 204 , 1951–1959 (2011).
McKay, B., Castellanos, M., Ebell, M., Whalen, C. C. & Handel, A. An attempt to reproduce a previous meta-analysis and a new analysis regarding the impact of directly observed therapy on tuberculosis treatment outcomes. PLoS ONE 14 , e0217219 (2019).
Manson, A. L. et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat. Genet. 49 , 395–402 (2017).
Dreyer, V. et al. High fluoroquinolone resistance proportions among multidrug-resistant tuberculosis driven by dominant L2 Mycobacterium tuberculosis clones in the Mumbai Metropolitan Region. Genome Med. 14 , 95 (2022).
Cox, H. et al. Potential contribution of HIV during first-line tuberculosis treatment to subsequent rifampicin-monoresistant tuberculosis and acquired tuberculosis drug resistance in South Africa: a retrospective molecular epidemiology study. Lancet Microbe 2 , e584–e593 (2021).
Wang, Z. et al. Epidemiological characteristics and risk factors of multidrug-resistant tuberculosis in Luoyang, China. Front. Public Health 11 , 1117101 (2023).
Hang, N. T. L. et al. Primary drug-resistant tuberculosis in Hanoi, Viet Nam: present status and risk factors. PLoS ONE 8 , e71867 (2013).
Vashakidze, L. et al. Prevalence and risk factors for drug resistance among hospitalized tuberculosis patients in Georgia. Int. J. Tuberc. Lung Dis. 13 , 1148–1153 (2009).
CAS PubMed Google Scholar
Andrews, J. R. et al. Predictors of multidrug-and extensively drug-resistant tuberculosis in a high HIV prevalence community. PLoS ONE 5 , e15735 (2010).
Mesfin, E. A. et al. Drug-resistance patterns of Mycobacterium tuberculosis strains and associated risk factors among multi drug-resistant tuberculosis suspected patients from Ethiopia. PLoS ONE 13 , e0197737 (2018).
Mbuh, T. P. et al. Predictors of drug-resistant tuberculosis among high-risk population diagnosed under national program conditions in the Littoral region, Cameroon. BioMed. Res. Int. 2021 , 8817442 (2021).
Skrahina, A. et al. Multidrug-resistant tuberculosis in Belarus: the size of the problem and associated risk factors. Bull. World Health Organ. 91 , 36–45 (2013).
Urrego, J. et al. The impact of ventilation and early diagnosis on tuberculosis transmission in Brazilian prisons. Am. J. Trop. Med. Hyg. 93 , 739–746 (2015).
Kerubo, G., Amukoye, E., Niemann, S. & Kariuki, S. Drug susceptibility profiles of pulmonary Mycobacterium tuberculosis isolates from patients in informal urban settlements in Nairobi, Kenya. BMC Infect. Dis. 16 , 583 (2016).
Oliveira, O. et al. Using Bayesian spatial models to map and to identify geographical hotspots of multidrug-resistant tuberculosis in Portugal between 2000 and 2016. Sci. Rep. 10 , 16646 (2020).
Jenkins, H. E. et al. Assessing spatial heterogeneity of multidrug-resistant tuberculosis in a high-burden country. Eur. Respir. J. 42 , 1291–1301 (2013).
Alene, K. A., Viney, K., McBryde, E. S. & Clements, A. C. A. Spatial patterns of multidrug resistant tuberculosis and relationships to socio-economic, demographic and household factors in northwest Ethiopia. PLoS ONE 12 , e0171800 (2017).
Paleckyte, A., Dissanayake, O., Mpagama, S., Lipman, M. C. & McHugh, T. D. Reducing the risk of tuberculosis transmission for HCWs in high incidence settings. Antimicrob. Resist. Infect. Control 10 , 106 (2021).
Escombe, A. R. et al. Tuberculosis transmission risk and infection control in a hospital emergency department in Lima, Peru. Int. J. Tuberc. Lung Dis. 14 , 1120–1126 (2010).
Telisinghe, L. et al. High tuberculosis prevalence in a South African prison: the need for routine tuberculosis screening. PLoS ONE 9 , e87262 (2014).
Dheda, K. et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 5 , 291–360 (2017).
Houben, R. M. G. J. & Glynn, J. R. A systematic review and meta-analysis of molecular epidemiological studies of tuberculosis: development of a new tool to aid interpretation. Trop. Med. Int. Health 14 , 892–909 (2009).
Chen, S. et al. Risk factors for multidrug resistance among previously treated patients with tuberculosis in eastern China: a case-control study. Int. J. Infect. Dis. 17 , e1116-20 (2013).
Pradipta, I. S., Forsman, L. D., Bruchfeld, J., Hak, E. & Alffenaar, J. W. Risk factors of multidrug-resistant tuberculosis: a global systematic review and meta-analysis. J. Infect. 77 , 469–478 (2018).
Lomtadze, N. et al. Prevalence and risk factors for multidrug-resistant tuberculosis in the Republic of Georgia: a population-based study. Int. J. Tuberc. Lung Dis. 13 , 68–73 (2009).
Lee, E. G. et al. Age-stratified anti-tuberculosis drug resistance profiles in South Korea: a multicenter retrospective study. BMC Infect. Dis. 20 , 446 (2020).
Behr, M. A., Edelstein, P. H. & Ramakrishnan, L. Revisiting the timetable of tuberculosis. BMJ 362 , k2738 (2018).
Oladimeji, O. et al. Gender and drug-resistant tuberculosis in Nigeria. Trop. Med. Infect. Dis. 8 , 104 (2023).
McQuaid, C. F., Horton, K. C., Dean, A. S., Knight, G. M. & White, R. G. The risk of multidrug-or rifampicin-resistance in males versus females with tuberculosis. Eur. Respir. J. 56 , 2000626 (2020).
O’Donnell, M. R. et al. Extensively drug-resistant tuberculosis in women, KwaZulu-Natal, South Africa. Emerg. Infect. Dis. 17 , 1942–1945 (2011).
Gandhi, N. R. et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368 , 1575–1580 (2006).
Kolla, B. P., Oesterle, T., Gold, M., Southwick, F. & Rummans, T. Infectious diseases occurring in the context of substance use disorders: a concise review. J. Neurol. Sci. 411 , 116719 (2020).
Soboka, M. et al. Substance use disorders and adherence to antituberculosis medications in Southwest Ethiopia: a prospective cohort study. BMJ Open 11 , e043050 (2021).
Mekonnen, H. S. & Azagew, A. W. Non-adherence to anti-tuberculosis treatment, reasons and associated factors among TB patients attending at Gondar town health centers, Northwest Ethiopia. BMC Res. Notes 11 , 691 (2018).
Chaves Torres, N. M., Quijano Rodríguez, J. J., Porras Andrade, P. S., Arriaga, M. B. & Netto, E. M. Factors predictive of the success of tuberculosis treatment: a systematic review with meta-analysis. PLoS ONE 14 , e0226507 (2019).
Dixit, A. et al. Modern lineages of Mycobacterium tuberculosis were recently introduced in Western India and demonstrate increased transmissibility. Preprint at medRxiv https://doi.org/10.1101/2022.01.04.22268645 (2022).
Casali, N. et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 22 , 735–745 (2012).
Gygli, S. M. et al. Publisher Correction: prisons as ecological drivers of fitness-compensated multidrug-resistant Mycobacterium tuberculosis . Nat. Med. 27 , 1308–1308 (2021).
Loiseau, C. et al. The relative transmission fitness of multidrug-resistant Mycobacterium tuberculosis in a drug resistance hotspot. Nat. Commun. 14 , 1988 (2023).
Gygli, S. M., Borrell, S., Trauner, A. & Gagneux, S. Antimicrobial resistance in Mycobacterium tuberculosis : mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 41 , 354–373 (2017).
Li, S. et al. CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis . Nat. Microbiol. 7 , 766–779 (2022).
Nguyen, L. & Pieters, J. Mycobacterial subversion of chemotherapeutic reagents and host defense tactics: challenges in tuberculosis drug development. Annu. Rev. Pharmacol. Toxicol. 49 , 427–453 (2009).
Morris, R. P. et al. Ancestral antibiotic resistance in Mycobacterium tuberculosis . Proc. Natl Acad. Sci. USA 102 , 12200–12205 (2005).
Mailaender, C. et al. The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis . Microbiology 150 , 853–864 (2004).
Rodriguez-Rivera, F. P., Zhou, X., Theriot, J. A. & Bertozzi, C. R. Visualization of mycobacterial membrane dynamics in live cells. J. Am. Chem. Soc. 139 , 3488–3495 (2017).
Madsen, C. T. et al. Methyltransferase Erm(37) slips on rRNA to confer atypical resistance in Mycobacterium tuberculosis . J. Biol. Chem. 280 , 38942–38947 (2005).
Wang, F., Cassidy, C. & Sacchettini, J. C. Crystal structure and activity studies of the Mycobacterium tuberculosis beta-lactamase reveal its critical role in resistance to β-lactam antibiotics. Antimicrob. Agents Chemother. 50 , 2762–2771 (2006).
Chambers, H. F., Kocagoz, T., Sipit, T., Turner, J. & Hopewell, P. C. Activity of amoxicillin/clavulanate in patients with tuberculosis. Clin. Infect. Dis. 26 , 874–877 (1998).
Donald, P. R. & Sirge, F. A. Early bactericidal activity of amoxicillin in combination with clavulanic acid in patients with sputum smear-positive pulmonary tuberculosis. Scand. J. Infect. Dis. 33 , 466–469 (2001).
Hugonnet, J.-E., Tremblay, L. W., Boshoff, H. I., Barry, C. E. & Blanchard, J. S. Meropenem-clavulanate is effective against extensively drug-desistant Mycobacterium tuberculosis . Science 323 , 1215–1218 (2009).
Vargas, R. Jr et al. Phase variation as a major mechanism of adaptation in Mycobacterium tuberculosis complex. Proc. Natl Acad. Sci. USA 120 , e2301394120 (2023).
Vargas, R. Jr Role of epistasis in amikacin, kanamycin, bedaquiline, and clofazimine resistance in Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 65 , e0116421 (2021).
Farhat, M. R. et al. Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am. J. Respir. Crit. Care Med. 194 , 621–630 (2016).
Nebenzahl-Guimaraes, H., Jacobson, K. R., Farhat, M. R. & Murray, M. B. Systematic review of allelic exchange experiments aimed at identifying mutations that confer drug resistance in Mycobacterium tuberculosis . J. Antimicrob. Chemother. 69 , 331–342 (2013).
Kadura, S. et al. Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J. Antimicrob. Chemother. 75 , 2031–2043 (2020).
Zhang, Y. & Yew, W. W. Mechanisms of drug resistance in Mycobacterium tuberculosis . Int. J. Tuberc. Lung Dis. 13 , 1320–1330 (2009).
Green, A. G. et al. Analysis of genome-wide mutational dependence in naturally evolving Mycobacterium tuberculosis populations. Mol. Biol. Evol. 40 , msad131 (2023).
Barilar, I. et al. Quantitative measurement of antibiotic resistance in Mycobacterium tuberculosis reveals genetic determinants of resistance and susceptibility in a target gene approach. Nat. Commun. 15 , 488 (2024).
Farhat, M. R. et al. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat. Commun. 10 , 2128 (2019).
Walker, T. M. et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe 3 , e265–e273 (2022).
Ghodousi, A. et al. Isoniazid resistance in Mycobacterium tuberculosis is a heterogeneous phenotype composed of overlapping MIC distributions with different underlying resistance mechanisms. Antimicrob. Agents Chemother. 63 , e00092-19 (2019).
Spitaleri, A., Ghodousi, A., Miotto, P. & Cirillo, D. M. Whole genome sequencing in Mycobacterium tuberculosis . Ann. Transl. Med. 7 , S197 (2019).
Farhat, M. R. et al. Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis . J. Clin. Microbiol. 54 , 727–733 (2016).
Chen, M. L. et al. Beyond multidrug resistance: leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction. EBioMedicine 43 , 356–369 (2019).
Green, A. G. et al. A convolutional neural network highlights mutations relevant to antimicrobial resistance in Mycobacterium tuberculosis . Nat. Commun. 13 , 3817 (2022).
Yang, Y. et al. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics 34 , 1666–1671 (2018).
Gröeschel, M. I. et al. GenTB: a user-friendly genome-based predictor for tuberculosis resistance powered by machine learning. Genome Med. 13 , 138 (2021).
Safi, H. et al. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc. Natl Acad. Sci. USA 116 , 19665–19674 (2019).
Hicks, N. D. et al. Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3 , 1032–1042 (2018).
Liu, Q. et al. Tuberculosis treatment failure associated with evolution of antibiotic resilience. Science 378 , 1111–1118 (2022).
Kreutzfeldt, K. M. et al. CinA mediates multidrug tolerance in Mycobacterium tuberculosis . Nat. Commun. 13 , 2203 (2022).
Martini, M. C. et al. Loss of RNase J leads to multi-drug tolerance and accumulation of highly structured mRNA fragments in Mycobacterium tuberculosis . PLoS Pathog. 18 , e1010705 (2022).
Andersson, D. I., Nicoloff, H. & Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 17 , 479–496 (2019).
Vargas, R. et al. In-host population dynamics of Mycobacterium tuberculosis complex during active disease. eLife 10 , e61805 (2021).
Nimmo, C. et al. Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine 55 , 102747 (2020).
Engelthaler, D. M. et al. Minority Mycobacterium tuberculosis genotypic populations as an indicator of subsequent phenotypic resistance. Am. J. Respir. Cell Mol. Biol. 61 , 789–791 (2019).
WHO Standard: Universal Access to Rapid Tuberculosis Diagnostics. World Health Organization https://www.who.int/publications/i/item/9789240071315 (2023).
Report of the 16th Meeting of the Strategic and Technical Advisory Group for Tuberculosis 2016. World Health Organization https://www.who.int/publications/m/item/report-of-the-16th-meeting-of-the-strategic-and-technical-advisory-group-for-tb (2016).
Jacobson, K. R. et al. Implications of failure to routinely diagnose resistance to second-line drugs in patients with rifampicin-resistant tuberculosis on Xpert MTB/RIF: a multisite observational study. Clin. Infect. Dis. 64 , 1502–1508 (2017).
Oga-Omenka, C. et al. Factors influencing diagnosis and treatment initiation for multidrug-resistant/rifampicin-resistant tuberculosis in six sub-Saharan African countries: a mixed-methods systematic review. BMJ Glob. Health 5 , e002280 (2020).
Svadzian, A., Sulis, G., Gore, G., Pai, M. & Denkinger, C. M. Differential yield of universal versus selective drug susceptibility testing of patients with tuberculosis in high-burden countries: a systematic review and meta-analysis. BMJ Glob. Health 5 , e003438 (2020).
Kim, S. J. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur. Respir. J. 25 , 564–569 (2005).
Technical Manual for Drug Susceptibility Testing of Medicines Used in the Treatment of Tuberculosis. World Health Organization https://www.who.int/publications/i/item/9789241514842 (2018).
Yu, H.-J. et al. Performance evaluation of the BACTEC MGIT 960 system for rifampin drug-susceptibility testing of Mycobacterium tuberculosis using the current WHO critical concentration. J. Clin. Microbiol. 61 , e01086-22 (2023).
Shea, J. et al. Low-level rifampin resistance and rpoB mutations in Mycobacterium tuberculosis : an analysis of whole-genome sequencing and drug susceptibility test data in New York. J. Clin. Microbiol. 59 , e01885-20 (2021).
Torrea, G. et al. Variable ability of rapid tests to detect Mycobacterium tuberculosis rpoB mutations conferring phenotypically occult rifampicin resistance. Sci. Rep. 9 , 11826 (2019).
Automated Real-time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB/RIF System: Policy Statement. World Health Organization https://www.who.int/publications/i/item/9789241501545 (2011).
Rapid Implementation of the Xpert MTB/RIF Diagnostic Test: Technical and Operational “How-to”; Practical Considerations. World Health Organization https://www.who.int/publications/i/item/9789241501569 (2011).
Dorman, S. E. et al. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect. Dis. 18 , 76–84 (2018).
Albert, H. et al. Development, roll-out and impact of Xpert MTB/RIF for tuberculosis: what lessons have we learnt and how can we do better? Eur. Respir. J. 48 , 516–525 (2016).
Zifodya, J. S. et al. Xpert Ultra versus Xpert MTB/RIF for pulmonary tuberculosis and rifampicin resistance in adults with presumptive pulmonary tuberculosis. Cochrane Database Syst. Rev. 2021 , CD009593 (2021).
Penn-Nicholson, A. et al. A prospective multicentre diagnostic accuracy study for the Truenat tuberculosis assays. Eur. Respir. J. 58 , 2100526 (2021).
Gomathi, N. S. et al. Validation of an indigenous assay for rapid molecular detection of rifampicin resistance in presumptive multidrug-resistant pulmonary tuberculosis patients. Indian J. Med. Res. 152 , 482–489 (2020).
Molbio Diagnostics: Molbio launches Truenat MTB-INH test for drug resistance in TB patients. Health News, ET HealthWorld https://health.economictimes.indiatimes.com/news/diagnostics/molbio-launches-truenat-mtb-inh-test-for-drug-resistance-in-tb-patients/96963161 (2023).
Theron, G. et al. Feasibility, accuracy, and clinical effect of point-of-care Xpert MTB/RIF testing for tuberculosis in primary-care settings in Africa: a multicentre, randomised, controlled trial. Lancet 383 , 424–435 (2014).
Yoon, C. et al. Impact of Xpert MTB/RIF testing on tuberculosis management and outcomes in hospitalized patients in Uganda. PLoS ONE 7 , e48599 (2012).
Di Tanna, G. L. et al. Effect of Xpert MTB/RIF on clinical outcomes in routine care settings: individual patient data meta-analysis. Lancet Glob. Health 7 , e191–e199 (2019).
Churchyard, G. J. et al. Xpert MTB/RIF versus sputum microscopy as the initial diagnostic test for tuberculosis: a cluster-randomised trial embedded in South African roll-out of Xpert MTB/RIF. Lancet Glob. Health 3 , 450–457 (2015).
Cattamanchi, A. et al. Multicomponent strategy with decentralized molecular testing for tuberculosis. N. Engl. J. Med. 385 , 2441–2450 (2021).
Penn-Nicholson, A. et al. Detection of isoniazid, fluoroquinolone, ethionamide, amikacin, kanamycin, and capreomycin resistance by the Xpert MTB/XDR assay: a cross-sectional multicentre diagnostic accuracy study. Lancet Infect. Dis. 22 , 242–249 (2022).
Cao, Y. et al. Xpert MTB/XDR: a 10-color reflex assay suitable for point-of-care settings to detect isoniazid, fluoroquinolone, and second-line-injectable-drug resistance directly from Mycobacterium tuberculosis-positive sputum. J. Clin. Microbiol. 59 , e02314-20 (2021).
De Vos, M. et al. Comparative analytical evaluation of four centralized platforms for the detection of Mycobacterium tuberculosis complex and resistance to rifampicin and isoniazid. J. Clin. Microbiol. 59 , e02168-20 (2021).
Meaza, A. et al. Evaluation of genotype MTBDRplus VER 2.0 line probe assay for the detection of MDR-TB in smear positive and negative sputum samples. BMC Infect. Dis. 17 , 280 (2017).
Nathavitharana, R. R. et al. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. Eur. Respir. J. 49 , 1601075 (2017).
Driesen, M. et al. Evaluation of a novel line probe assay to detect resistance to pyrazinamide, a key drug used for tuberculosis treatment. Clin. Microbiol. Infect. 24 , 60–64 (2018).
Willby, M. J. et al. Detection of Mycobacterium tuberculosis pncA mutations by the Nipro Genoscholar PZA-TB II assay compared to conventional sequencing. Antimicrob. Agents Chemother. 62 , e01871-17 (2018).
Catalogue of Mutations in Mycobacterium tuberculosis Complex and their Association with Drug Resistance, 2nd edn. World Health Organization https://www.who.int/publications/i/item/9789240082410 (2023).
Ismail, N. et al. Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis : a systematic review and individual isolate data analysis. Lancet Microbe 2 , e604–e616 (2021).
An, Q., Lin, R., Yang, Q., Wang, C. & Wang, D. Evaluation of genetic mutations associated with phenotypic resistance to fluoroquinolones, bedaquiline, and linezolid in clinical Mycobacterium tuberculosis : a systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 34 , 214–226 (2023).
Gan, W. C., Ng, H. F. & Ngeow, Y. F. Mechanisms of linezolid resistance in mycobacteria. Pharmaceuticals 16 , 784 (2023).
CRyPTIC Consortium & The 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379 , 1403–1415 (2018).
Pankhurst, L. J. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir. Med. 4 , 49–58 (2016).
Use of Targeted Next-generation Sequencing to Detect Drug-resistant Tuberculosis: Rapid Communication. World Health Organization https://www.who.int/publications/i/item/9789240076372 (2023).
The Use of Next-Generation Sequencing for the Surveillance of Drug-Resistant Tuberculosis: An Implementation Manual. World Health Organization https://www.who.int/publications/i/item/9789240078079 (2023).
Dippenaar, A. et al. Nanopore sequencing for Mycobacterium tuberculosis : a critical review of the literature, new developments, and future opportunities. J. Clin. Microbiol. 60 , e00646-21 (2022).
Sanchez-Padilla, E. et al. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N. Engl. J. Med. 372 , 1181–1182 (2015).
Ng, K. C. et al. Xpert Ultra can unambiguously identify specific rifampin resistance-conferring mutations. J. Clin. Microbiol. 56 , 10–1128 (2018).
Daum, L. T. et al. Next-generation sequencing for characterizing drug resistance-conferring Mycobacterium tuberculosis genes from clinical isolates in the Ukraine. J. Clin. Microbiol. 56 , e00009-18 (2018).
Tagliani, E. et al. Culture and next-generation sequencing-based drug susceptibility testing unveil high levels of drug-resistant-TB in Djibouti: results from the first national survey. Sci. Rep. 7 , 17672 (2017).
Walker, T. M. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13 , 137–146 (2013).
Mahomed, S., Mlisana, K., Cele, L. & Naidoo, K. Discordant line probe genotypic testing vs culture-based drug susceptibility phenotypic testing in TB endemic KwaZulu-Natal: impact on bedside clinical decision making. J. Clin. Tuberc. Mycobact. Dis. 20 , 100176 (2020).
Milimo, D. et al. Diagnosis of rifampicin-resistant tuberculosis: discordant results by diagnostic methods. Afr. J. Lab. Med. 7 , 1–4 (2018).
Votintseva, A. A. et al. Same-day diagnostic and surveillance data for ttuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 55 , 1285–1298 (2017).
Brown, A. C. et al. Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J. Clin. Microbiol. 53 , 2230–2237 (2015).
Doyle, R. M. et al. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J. Clin. Microbiol. 56 , e00666–18 (2018).
Goig, G. A. et al. Whole-genome sequencing of Mycobacterium tuberculosis directly from clinical samples for high-resolution genomic epidemiology and drug resistance surveillance: an observational study. Lancet Microbe 1 , e175–e183 (2020).
Van Rie, A. et al. Sequencing mycobacteria and algorithm-determined resistant tuberculosis treatment (SMARTT): a study protocol for a phase-IV pragmatic randomized controlled patient management strategy trial. Trials 23 , 864 (2022).
Sibandze, D. B. et al. Rapid molecular diagnostics of tuberculosis resistance by targeted stool sequencing. Genome Med. 14 , 52 (2022).
Iyer, A. et al. Operationalising targeted next-generation sequencing for routine diagnosis of drug-resistant TB. Public Health Action. 13 , 43–49 (2023).
Cox, H. et al. Whole-genome sequencing has the potential to improve treatment for rifampicin-resistant tuberculosis in high-burden settings: a retrospective cohort study. J. Clin. Microbiol. 60 , e02362–21 (2022).
Kwong, J. C., McCallum, N., Sintchenko, V. & Howden, B. P. Whole genome sequencing in clinical and public health microbiology. Pathology 47 , 199–210 (2015).
Conradie, F. et al. Treatment of highly drug-resistant pulmonary tuberculosis. N. Engl. J. Med. 382 , 893–902 (2020).
Nyang’wa, B.-T. et al. A 24-week, all-oral regimen for rifampin-resistant tuberculosis. N. Engl. J. Med. 387 , 2331–2343 (2022).
Padmapriyadarsini, C. et al. Bedaquiline, delamanid, linezolid and clofazimine for treatment of pre-extensively drug-resistant tuberculosis. Clin. Infect. Dis. 76 , e938–e946 (2022).
Pym, A. S. et al. Bedaquiline in the treatment of multidrug- and extensively drug-resistant tuberculosis. Eur. Respir. J. 47 , 564–574 (2016).
Schnippel, K. et al. Effect of bedaquiline on mortality in South African patients with drug-resistant tuberculosis: a retrospective cohort study. Lancet Respir. Med. 6 , 699–706 (2018).
Ndjeka, N. et al. High treatment success rate for multidrug-resistant and extensively drug-resistant tuberculosis using a bedaquiline-containing treatment regimen. Eur. Respir. J. 52 , 1801528 (2018).
Ndjeka, N. et al. Treatment outcomes 24 months after initiating short bedaquiline-containing or injectable-containing rifampicin-resistant tuberculosis treatment regimens: a retrospective cohort study in South Africa. Lancet Infect. Dis. 22 , 1042–1051 (2022).
Zhao, Y. et al. Improved treatment outcomes with bedaquiline when substituted for second-line injectable agents in multidrug-resistant tuberculosis: a retrospective cohort study. Clin. Infect. Dis. 68 , 1522–1529 (2019).
Conradie, F. et al. Bedaquiline–pretomanid–linezolid regimens for drug-resistant tuberculosis. N. Engl. J. Med. 387 , 810–823 (2022).
Esmail, A. et al. An all-oral 6-month regimen for multidrug-resistant tuberculosis: a multicenter, randomized controlled clinical trial (the NExT study). Am. J. Respir. Crit. Care Med. 205 , 1214–1227 (2022).
Goodall, R. L. et al. Evaluation of two short standardised regimens for the treatment of rifampicin-resistant tuberculosis (STREAM stage 2): an open-label, multicentre, randomised, non-inferiority trial. Lancet 400 , 1858–1868 (2022).
WHO Operational Handbook on Tuberculosis. Module 4: Treatment of Drug-resistant Tuberculosis. World Health Organization https://www.who.int/publications/i/item/9789240006997 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02589782 (2021).
Clinical Trial Results Offer Hope to DR-TB patients with short, effective treatment. Médecins Sans Frontières https://www.msf.org/clinical-trial-finds-short-effective-safe-DR-TB-treatment (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02333799 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03086486 (2023).
Drug-resistant Tuberculosis Clinical Trials Progress Report. RESIST-TB https://www.resisttb.org/clinical-trials-progress-report (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02619994 (2019).
Mok, J. et al. 9 months of delamanid, linezolid, levofloxacin, and pyrazinamide versus conventional therapy for treatment of fluoroquinolone-sensitive multidrug-resistant tuberculosis (MDR-END): a multicentre, randomised, open-label phase 2/3 non-inferiority trial in South Korea. Lancet 400 , 1522–1530 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04062201 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02754765 (2023).
Guglielmetti, L. et al. Evaluating newly approved drugs for multidrug-resistant tuberculosis (endTB): study protocol for an adaptive, multi-country randomized controlled trial. Trials 22 , 651 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03896685 (2022).
ShORRT Initiative. Tropical Disease Research https://tdr.who.int/activities/shorrt-research-package (2022).
Tuberculosis Treatment: 2023 Pipeline Report. Treatment Action Group https://www.treatmentactiongroup.org/resources/pipeline-report/2023-pipeline-report/ (2023).
WHO Operational Handbook on Tuberculosis. Module 4: Treatment — Drug-resistant Tuberculosis Treatment, 2022 Update. World Health Organization https://www.who.int/publications/i/item/9789240065116 (2022).
Maugans, C. et al. Best practices for the care of pregnant people living with TB. Int. J. Tuberc. Lung Dis. 27 , 357–366 (2023).
Patankar, S. et al. Making the case for all-oral, shorter regimens for children with drug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 208 , 130–131 (2023).
Turkova, A. et al. Shorter treatment for nonsevere tuberculosis in African and Indian children. N. Engl. J. Med. 386 , 911–922 (2022).
Management of Multidrug-resistant Tuberculosis in Children: A Field Guide. Sentinel Project on Pediatric Drug-Resistant Tuberculosis https://sentinel-project.org/wp-content/uploads/2022/03/DRTB-Field-Guide-2021_v5.pdf (2022).
Furin, J., Tommasi, M. & Garcia-Prats, A. J. Drug-resistant tuberculosis: will grand promises fail children and adolescents? Lancet Child. Adolesc. Health 2 , 237–238 (2018).
Karo, B. et al. Isoniazid (INH) mono-resistance and tuberculosis (TB) treatment success: analysis of European surveillance data, 2002 to 2014. Eur. Surveill. 24 , 1800392 (2019).
Gegia, M., Winters, N., Benedetti, A., Soolingen, D. & Menzies, D. Treatment of isoniazid-resistant tuberculosis with first-line drugs: a systematic review and meta-analysis. Lancet Infect. Dis. 17 , 223–234 (2017).
Furin, J., Cox, H. & Pai, M. Tuberculosis. Lancet 393 , 1642–1656 (2019).
WHO Consolidated Guidelines on Tuberculosis: Module 1: Prevention: Tuberculosis Preventive Treatment. World Health Organization https://www.who.int/publications/i/item/9789240001503 (2020).
Guidelines for Programmatic Management of Tuberculosis Preventive Treatment in India. National TB Elimination Programme https://tbcindia.gov.in/WriteReadData/l892s/Guidelines%20for%20Programmatic%20Management%20of%20Tuberculosis%20Preventive%20Treatment%20in%20India.pdf (2021).
Latent Tuberculosis Infection: Updated and Consolidated Guidelines for Programmatic Management. World Health Organization https://www.who.int/publications/i/item/9789241550239 (2018).
Kherabi, Y., Tunesi, S., Kay, A. & Guglielmetti, L. Preventive therapy for contacts of drug-resistant tuberculosis. Pathogens 11 , 1189 (2022).
Huang, C. C. et al. Isoniazid preventive therapy in contacts of multidrug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 202 , 1159–1168 (2020).
Seddon, J. A. et al. Levofloxacin versus placebo for the prevention of tuberculosis disease in child contacts of multidrug-resistant tuberculosis: study protocol for a phase-III cluster randomised controlled trial (TB-CHAMP). Trials 19 , 693 (2018).
Fox, G. J. et al. Levofloxacin versus placebo for the treatment of latent tuberculosis among contacts of patients with multidrug-resistant tuberculosis (the VQUIN MDR trial): a protocol for a randomised controlled trial. BMJ Open 10 , e033945 (2020).
First Effective Treatment to Prevent Multidrug-Resistant TB. UCL News https://www.ucl.ac.uk/news/2023/nov/first-effective-treatment-prevent-multidrug-resistant-tb (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03568383 (2023).
WHO announces forthcoming updates on tuberculosis preventive treatment. World Health Organization https://www.who.int/news/item/13-02-2024-who-announces-forthcoming-updates-on-tuberculosis-preventive-treatment (2024).
Zimmerman, E., Smith, J., Banay, R., Kau, M. & Garfin, A. M. C. G. Behavioural barriers and perceived trade-offs to care-seeking for tuberculosis in the Philippines. Glob. Public Health 17 , 210–222 (2022).
Daftary, A., Frick, M., Venkatesan, N., & Pai, M. Fighting TB stigma: we need to apply lessons learnt from HIV activism. BMJ Glob. Health 2 , e000515 (2017).
Pradhan, A. et al. Internalized and perceived stigma and depression in pulmonary tuberculosis: do they explain the relationship between drug sensitivity status and adherence? Front. Psychiatry 13 , 869647 (2022).
Redwood, L. et al. Depression, stigma and quality of life in people with drug-susceptible TB and drug-resistant TB in Vietnam. Int. J. Tuberc. Lung Dis. 25 , 461–467 (2021).
Daftary, A., Padayatchi, N. & O’Donnell, M. Preferential adherence to antiretroviral therapy over tuberculosis treatment: a qualitative study of drug-resistant TB/HIV co-infected patients in South Africa. Glob. Public Health 9 , 1107–1116 (2014).
Susanto, T. D. et al. Anxiety and depression level of patients with multidrug-resistant tuberculosis (MDR-TB) in two hospitals in Banten province, Indonesia. Dialogues Health 2 , 100115 (2023).
Walker, I. et al. Depression and anxiety in patients with multidrug-resistant tuberculosis in Nepal: an observational study. Public Health Action 9 , 42–48 (2019).
Sommerland, N. et al. Evidence-based interventions to reduce tuberculosis stigma: a systematic review. Int. J. Tuberc. Lung Dis. 21 , S81–S86 (2017).
As’hab, P. P., Keliat, B. A. & Wardani, I. Y. The effects of acceptance and commitment therapy on psychosocial impact and adherence of multidrug-resistant tuberculosis patients. J. Public Health Res. 11 , 2737 (2022).
Thomas, B. E. et al. Psycho-socio-economic issues challenging multidrug resistant tuberculosis patients: a systematic review. PLoS ONE 11 , e0147397 (2016).
Udwadia, Z. & Furin, J. Quality of drug-resistant tuberculosis care: gaps and solutions. J. Clin. Tuberc. Mycobact. Dis. 16 , 100101 (2019).
Pai, M., Dewan, P. K. & Swaminathan, S. Transforming tuberculosis diagnosis. Nat. Microbiol. 8 , 756–759 (2023).
McKenna, L. et al. The 1/4/6x24 campaign to cure tuberculosis quickly. Nat. Med. 11 , 2337 (2023).
Zaunbrecher, M. A., Sikes, R. D. Jr, Metchock, B., Shinnick, T. M. & Posey, J. E. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis . Proc. Natl Acad. Sci. USA 106 , 20004–20009 (2009).
Miotto, P. et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis . Eur. Respir. J. 50 , 1701354 (2017).
Rifat, D. et al. Mutations in fbiD (Rv2983) as a novel determinant of resistance to pretomanid and delamanid in Mycobacterium tuberculosis . Antimicrob. Agents Chemother. 65 , e01948–20 (2020).
Plinke, C. et al. embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J. Antimicrob. Chemother. 65 , 1359–1367 (2010).
Safi, H. et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 45 , 1190–1197 (2013).
Srivastava, S., Ayyagari, A., Dhole, T. N., Nyati, K. K. & Dwivedi, S. K. emb nucleotide polymorphisms and the role of embB306 mutations in Mycobacterium tuberculosis resistance to ethambutol. Int. J. Med. Microbiol. 299 , 269–280 (2009).
Grant, S. S. et al. Baeyer-Villiger monooxygenases EthA and MymA are required for activation of replicating and non-replicating Mycobacterium tuberculosis inhibitors. Cell Chem. Biol. 23 , 666–677 (2016).
Dover, L. G. et al. EthA, a common activator of thiocarbamide-containing drugs acting on different mycobacterial targets. Antimicrob. Agents Chemother. 51 , 1055–1063 (2007).
Vilchèze, C. et al. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis . Mol. Microbiol. 69 , 1316–1329 (2008).
Zhang, Y., Dhandayuthapani, S. & Deretic, V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc. Natl Acad. Sci. USA 93 , 13212–13216 (1996).
Ando, H., Miyoshi-Akiyama, T., Watanabe, S. & Kirikae, T. A silent mutation in mabA confers isoniazid resistance on Mycobacterium tuberculosis . Mol. Microbiol. 91 , 538–547 (2014).
Vilchèze, C. et al. Altered NADH/NAD + ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 49 , 708–720 (2005).
Reeves, A. Z. et al. Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7 . Antimicrob. Agents Chemother. 57 , 1857–1865 (2013).
Liu, J. et al. Mutations in efflux pump Rv1258c (tap) cause resistance to pyrazinamide, isoniazid, and streptomycin in Mycobacterium tuberculosis . Front. Microbiol. 10 , 216 (2019).
Gopal, P. et al. Pyrazinamide triggers degradation of its target aspartate decarboxylase. Nat. Commun. 11 , 1661 (2020).
Scorpio, A. & Zhang, Y. Mutations in pncA , a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2 , 662–667 (1996).
Wong, S. Y. et al. Functional role of methylation of G518 of the 16S rRNA 530 loop by GidB in Mycobacterium tuberculosis . Antimicrob. Agents Chemother. 57 , 6311–6318 (2013).
Catalogue of Mutations in Mycobacterium tuberculosis Complex and their Association with Drug Resistance. World Health Organization https://www.who.int/publications/i/item/9789240028173 (2021).
Farhat, M. R. et al. Rifampicin and rifabutin resistance in 1003 Mycobacterium tuberculosis clinical isolates. J. Antimicrob. Chemother. 74 , 1477–1483 (2019).
Nahid, P. et al. Treatment of drug-resistant tuberculosis. An Official ATS/CDC/ERS/IDSA clinical practice guideline. Am. J. Respir. Crit. Care Med. 200 , e93–e142 (2019).
Download references
Author information
These authors contributed equally: Helen Cox, Marwan Ghanem.
Authors and Affiliations
Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA
Maha Farhat & Marwan Ghanem
Division of Pulmonary and Critical Care Medicine, Massachusetts General Hospital, Boston, MA, USA
Maha Farhat
Institute of Infectious Disease and Molecular Medicine, Wellcome Centre for Infectious Disease Research and Division of Medical Microbiology, University of Cape Town, Cape Town, South Africa
Division of Infectious Disease and Tropical Medicine, Heidelberg University Hospital, Heidelberg, Germany
Claudia M. Denkinger & Mirna S. Abd El Aziz
German Center for Infection Research (DZIF), partner site Heidelberg University Hospital, Heidelberg, Germany
Claudia M. Denkinger
P.D. Hinduja Hospital and Medical Research Centre, Mumbai, India
Camilla Rodrigues
Ulaanbaatar, Mongolia
Handaa Enkh-Amgalan
National TB Control Programme, Manzini, Eswatini
Debrah Vambe
School of Public and Population Health, University of Texas Medical Branch, Galveston, TX, USA
Cesar Ugarte-Gil
Department of Global Health and Social Medicine, Harvard Medical School, Boston, MA, USA
Jennifer Furin
McGill International TB Centre, McGill University, Montreal, Quebec, Canada
Madhukar Pai
You can also search for this author in PubMed Google Scholar
Contributions
M.P., M.F., H.C., C.M.D., J.F. and M.G. wrote the article and, together with C.R. and M.S.A.E.A. researched data for the article. All authors contributed substantially to the discussion of the content and reviewed and/or edited the manuscript before submission.
Corresponding author
Correspondence to Madhukar Pai .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Peer review
Peer review information.
Nature Reviews Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Clinicaltrials.gov: https://clinicaltrials.gov/
Supplementary information
Supplementary information.
Refers to the delivery of anti-tuberculosis drug treatment under direct observation of health workers, community workers or family members with the goal of improving adherence.
(XDR-TB). Defined as multidrug-resistant or rifampicin-resistant tuberculosis with further resistance to fluoroquinolones and to either bedaquiline or linezolid or both (key second-line drugs).
(Hr-TB). Defined as resistance to isoniazid and susceptibility to rifampicin.
(MDR-TB). Defined as resistance to rifampicin and isoniazid, the two most important first-line drugs used to treat tuberculosis (TB), regardless of resistance to other TB drugs.
Defined as multidrug-resistant or rifampicin-resistant tuberculosis with resistance to fluoroquinolones.
(RMR-TB). Defined as resistance to rifampicin (Rif), with susceptibility to isoniazid.
(RR-TB). Defined as resistance to rifampicin (Rif), regardless of resistance to other tuberculosis (TB) drugs. Individuals with RR-TB are treated with regimens similar to those for multidrug-resistant TB (MDR-TB) and are therefore grouped with MDR-TB as MDR/RR-TB.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Cite this article.
Farhat, M., Cox, H., Ghanem, M. et al. Drug-resistant tuberculosis: a persistent global health concern. Nat Rev Microbiol (2024). https://doi.org/10.1038/s41579-024-01025-1
Download citation
Accepted : 12 February 2024
Published : 22 March 2024
DOI : https://doi.org/10.1038/s41579-024-01025-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Microbiology newsletter — what matters in microbiology research, free to your inbox weekly.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Front Cell Infect Microbiol
Evolution of Mycobacterium tuberculosis drug resistance in the genomic era
Camus nimmo.
1 Systems Chemical Biology of Infection and Resistance Laboratory, Francis Crick Institute, London, United Kingdom
James Millard
2 Institute of Infection and Global Health, University of Liverpool, Liverpool, United Kingdom
Valwynne Faulkner
Johana monteserin, hannah pugh, eachan oliver johnson.
Mycobacterium tuberculosis has acquired drug resistance to all drugs that have been used against it, including those only recently introduced into clinical practice. Compared to other bacteria, it has a well conserved genome due to its role as an obligate human pathogen that has adapted to a niche over five to ten thousand years. These features facilitate reconstruction and dating of M. tuberculosis phylogenies, giving key insights into how resistance has been acquired and spread globally. Resistance to each new drug has occurred within five to ten years of clinical use and has occurred even more rapidly with recently introduced drugs. In most cases, resistance-conferring mutations come with a fitness cost, but this can be overcome by compensatory mutations which restore fitness to that of wild-type bacteria. It is likely that M. tuberculosis acquires drug resistance while maintaining limited genomic variability due the generation of low frequency within-host variation, combined with ongoing purifying selection causing loss of variants without a clear fitness advantage. However, variants that do confer an advantage, such as drug resistance, can increase in prevalence amongst all bacteria within a host and become the dominant clone. These resistant strains can then be transmitted leading to primary drug resistant infection in a new host. As many countries move towards genomic methods for diagnosis of M. tuberculosis infection and drug resistance, it is important to be aware of the implications for the evolution of resistance. Currently, understanding of resistance-conferring mutations is incomplete, and some targeted genetic diagnostics create their own selective pressures. We discuss an example where a rifampicin resistance-conferring mutation which was not routinely covered by standard testing became dominant. Finally, resistance to new drugs such as bedaquiline and delamanid is caused by individually rare mutations occurring across a large mutational genomic target that have been detected over a short time, and do not provide statistical power for genotype-phenotype correlation – in contrast to longer-established drugs that form the backbone of drug-sensitive antituberculosis therapy. Therefore, we need a different approach to identify resistance-conferring mutations of new drugs before their resistance becomes widespread, abrogating their usefulness.
Introduction
Mycobacterium tuberculosis is an ancient bacterial pathogen that has acquired drug resistance to all drugs that have been used against it, despite lacking several key mechanisms available to other bacteria to facilitate rapid spread of resistance such as horizontal gene transfer and mobile resistance elements. In the absence of such mechanisms, all antituberculosis drug resistance is conferred by genomic mutations, mostly single nucleotide polymorphisms (SNPs), that are propagated through replication of resistant bacteria and onward transmission. In this review, we discuss how M. tuberculosis can develop drug resistance despite maintaining a comparatively well-conserved genome compared to other bacterial pathogens ( Eldholm and Balloux, 2016 ) and give examples of how it has acquired resistance at the between-host and within-host levels. Finally, we assess how diagnostics may be affected by resistance and shape its emergence, and we outline the implications for identifying resistance to new drugs against tuberculosis that are entering clinical use.
Key features of the Mycobacterium tuberculosis genome
The whole genome sequence of H37Rv, originally isolated from a patient treated in New York in 1905 and now the most used laboratory strain of M. tuberculosis , was published in 1998 ( Cole et al., 1998 ). The most recent annotation reports it as 4.4 megabases in length, making it 33% smaller than Mycobacterium smegmatis [also named Mycolicibacterium smegmatis , ( Gupta et al., 2018 ), although the usefulness of this is contested ( Tortoli et al., 2019 )], and one of the smallest apart from Mycobacterium leprae (1.6 megabases). It contains 3906 coding genes, of which a large number are responsible for fatty acid metabolism due to the complex mycobacterial cell wall. It is very rich in guanine and cytosine residues, and unlike many other bacteria (e.g. gram-negatives) there is no evidence of recombination and no accessory genome ( Eldholm and Balloux, 2016 ). About 10% of the genome is devoted to a characteristic set of proline (P)- and glutamate (E)-rich proteins called the PE and PPE gene families, which are heterogenous and consist of numerous tandem repeats and are hypothesised to be surface antigens that are responsible for interaction with the host immune system ( Fishbein et al., 2015 ). They are difficult to resolve by short read sequencing and as a result have been historically excluded from many genomic analyses of M. tuberculosis .
M. tuberculosis has traditionally been viewed as a genetically homogenous bacterium that has evolved into a specialised human pathogen with a lower mutation rate than most other bacteria at 0.3 to 0.5 SNPs per genome per year ( Eldholm and Balloux, 2016 ). The M. tuberculosis complex (MTBC) is likely to have originated from the transition of an environmental mycobacterial ancestor shared with the pathogen M. canetti ( Soolingen et al., 1997 ). The transition came with a corresponding reduction in genome size and loss of the ability for genetic recombination or gene transfer, perhaps because it developed into a specialised pathogen that lives only in one ecological niche. The original divergence of the MTBC from environmental mycobacteria is likely to have happened in Africa and then been spread globally by human migration ( Gagneux, 2018 ). Animal-adapted strains of the MTBC, including M. bovis (cows) and M. caprae (goats) are likely to have been transferred from humans as evidenced by comparative genomic studies that show loss of genes from M. tuberculosis sensu stricto to other members of the MTBC ( Comas et al., 2013 ). Genetic evidence suggests that MTBC is likely to have originated around 5,000-10,000 years ago ( Bos et al., 2014 ; Kay et al., 2015 ; Chiner-Oms et al., 2019 ), corresponding with archaeological evidence of M. tuberculosis DNA and lipids in skeletal remains from 9,000 years ago ( Hershkovitz et al., 2008 ).
The modern MTBC comprises seven human-adapted lineages, which are phylogenetically distinct groups clades that have evolved separately, having diverged over a period of 500 to 3000 years ( O’Neill et al., 2019 ) and several animal-adapted strains. Lineages 1, 2, 3, 4 and 7 are traditionally referred to as M. tuberculosis sensu stricto , while lineages 5 and 6 are known as M. africanum . Lineages 5 and 6 are restricted to West Africa and lineage 7 to East Africa, suggesting that they may have specifically adapted to their host populations ( Asante-Poku et al., 2015 ). Lineage 1 to 4 are globally distributed, with lineages 2 and 4 being the most prevalent worldwide ( Gagneux, 2018 )
Mechanisms of drug resistance
The majority of M. tuberculosis antibiotic resistance is conferred by genomic mutations – usually SNPs or small insertions or deletions, and occasionally larger deletions or inversions. Given the lack of horizontal gene transfer or episomal resistance genes ( Boritsch et al., 2016 ), these generally arise spontaneously and are chromosomally encoded, with spread through replication within host and onward transmission between hosts of resistant bacteria.
In contrast to organisms which exhibit horizontal gene transfer and therefore can also acquire extrachromosomal drug-inactivating resistance genes, there are three main mechanisms through which antituberculosis drug resistance can be acquired: target-based mutations, activator mutations and modulation of efflux pumps. Target-based mutations are where the drug target itself becomes mutated, usually preventing drug binding. Many antituberculosis drugs are administered as prodrugs that require activation by bacterial enzymes to produce their active form. In these cases, mutations of drug activators can lead to resistance. Finally, efflux pumps may pump active drug out of the bacterial cell, although there are fewer examples of these. Examples of each of these mechanisms are shown in Table 1 ( Silva and Palomino, 2011 ; Dookie et al., 2018 ). Some drugs have multiple mechanisms of resistance, for example isoniazid resistance can be conferred by target-based ( inhA ) or activator ( katG ) mutations. Some mutations may lead to cross-resistance, while others monoresistance. For example, atpE is the target for only bedaquiline. However, resistance to bedaquiline, clofazimine, and even new tuberculosis inhibitors like BRD-9327 can be conferred by efflux pump regulator mutations in Rv0678 ( Johnson et al., 2020 ).
Table 1
Categories of mutation leading to M. tuberculosis drug resistance.
| Category | Gene (drug) | Mechanism of resistance |
|---|---|---|
| Target-based | (rifampicin) | Rifampicin is unable to bind RNA polymerase, responsible for mRNA elongation ( ) |
| (isoniazid, ethionamide) | Both drugs unable to bind NADH-dependent enoyl-acyl carrier protein reductase responsible for mycolic acid synthesis ( ) | |
| (fluoroquinolones) | Mutations prevent binding to DNA gyrase required for DNA replication ( ) | |
| (aminoglycosides) | Prevent binding to 23S ribosomal RNA which prevents protein synthesis ( ) | |
| (bedaquiline) | Prevents binding to F1F0 proton ATP synthase, part of electron transport chain ( ) | |
| (ethambutol) | Mutations in the mycobacterial arabinosyl transferase enzyme preventing synthesis of arabinogalactan for the cell wall ( ) | |
| Drug activator | (isoniazid) | Isoniazid is activated by the katG-encoded catalase-peroxidase enzyme. S315T mutations prevent activation while maintaining native gene function ( ) |
| (ethionamide) | Mutations in the activating mono-oxygenase enzyme encoded by ethA, or its regulator ethR ( ) | |
| (pyrazinamide) | Diverse range of mutations in pyrazinamide activating PZase encoded by pncA lead to resistance, including any loss of function mutation as PZase loss does not impair fitness ( ) | |
| , fgd1, ddn (delamanid/pretomanid) | Wide variety of mutations inactivating enzymes ddn and co-enzyme fgd1. Also mutations in synthetic pathway for F420 cofactor required for activation (fbiA/B/C) ( ) | |
| Efflux pumps | (bedaquiline, clofazimine) | Mutations affecting or preventing function of the Rv0678 repressor of the MmpL5 efflux pump lead to overexpression of the pump and presumed efflux of bedaquiline and clofazimine ( ) |
Factors affecting acquisition of drug resistance
Prevalence of drug resistance varies by drug, patterns of drug usage (including the combinations of drugs it was used with), bacterial genetic background and country ( Brynildsrud et al., 2018 ; Ektefaie et al., 2021 ). The ability of M. tuberculosis to acquire drug resistance to each drug is underpinned by the rate at which spontaneous mutants arise and survive. This is different for each drug, with pyrazinamide having a particularly high rate of resistance acquisition in vitro and rifampicin a lower rate ( Figure 1 ) ( McGrath et al., 2014 ; David, 1970 ; Billington et al., 1999 ; Werngren abd Hoffner, 2003 ; O'Sullivan et al., 2008 ; Bergval et al., 2009, 2012 ; Huitric et al., 2010 ; Kana et al., 2010 ; Ford et al., 2011 ; Stoffels et al., 2012 ). However, the clinical relevance of in vitro mutation rates is only one aspect of the likely robustness of a drug against the development of resistance against it. For example, the studies examining spontaneous development of bedaquiline-resistant mutants ( Huitric et al., 2010 ) recorded the rate at which bacteria developed atpE mutations, which is the main gene determining bedaquiline resistance in vitro but not in vivo , where virtually all clinically reports mutations are in the Rv0678 gene ( Huitric et al., 2010 ). This is likely because atpE is an essential gene and mutations carry a high fitness cost in vitro , underlining the importance of understanding in vivo fitness costs of mutations.

Spontaneous in vitro mutation rates for key antituberculous drugs ( McGrath et al., 2014 ). Rates were measured in a variety of laboratory and clinical strains from varying bacterial lineages. Drug names abbreviated: RIF, rifampicin; INH, isoniazid; PZA, pyrazinamide; EMB, ethambutol; BDQ, bedaquiline; STR, streptomycin; DCS, D-cycloserine. Concentrations in mg/L shown in brackets followed by original reference ( David, 1970 ; Billington et al., 1999 ; Werngren and Hoffner, 2003 ; O’Sullivan et al., 2008 ; Bergval et al., 2009 , 2012 ; Huitric et al., 2010 ; Kana et al., 2010 ; Ford et al., 2011 ; Stoffels et al., 2012 ).
Bacterial genetic background is also likely to affect the ability of certain strains to acquire drug resistance. The best described example of this is higher prevalence of drug resistance amongst lineage 2 strains compared to other bacterial lineages ( Parwati et al., 2010 ; Ektefaie et al., 2021 ). It is likely that lineage 2 M. tuberculosis strains have a greater inherent ability to acquire drug resistance, and this has now been suggested by multiple studies ( Ford et al., 2013 ; Hakamata et al., 2020 ; Nimmo et al., 2020 ; Ortiz et al., 2021 ), although at least one study did not find this link ( Guerra-Assunção et al., 2015 ). While the mechanisms through which this may occur have not been fully elucidated, in vitro work with M. smegmatis (a related mycobacterium often used for laboratory studies) has shown that ribosomal mutations can lead to resistance to multiple antibiotics and enhanced bacterial survival ( Gomez et al., 2017 ). An alternative explanation for higher rates of drug resistance amongst lineage 2 strains may be the founder effect, where lineage 2 strains that were already drug resistant clonally expanded rapidly in an area with high rates of transmission ( Grandjean et al., 2015 ).
Finally, country-specific factors have been shown to influence the development of resistance even within a given bacterial strain. One example from a reconstructed phylogeny of the Central Asian Clade, a subgroup of lineage 2.2 (Beijing strain), showed it was in circulation in former Soviet republics in the 1960s and 1970s, before its introduction into Afghanistan in the 1980s ( Eldholm et al., 2016 ). Many resistance mutations arose independently amongst strains that were circulating in the former Soviet republics, while very few apart from the original lineage-defining rpoB mutation were present in the Afghan strains, with the vast majority of these mutations arising in the years after the collapse of the Soviet Union. Another analysis of the global spread of lineage 4 found that it was likely to have been dispersed from Europe during colonial expansion, that most drug resistance conferring mutations arose and were subsequently spread within individual countries ( Brynildsrud et al., 2018 ). Taken together, this suggests that a variety of factors that are hard to quantify, such as differing healthcare systems and political instability that are likely to impact on patterns of antimicrobial prescription, supply and usage. Additionally, country-level variation in sequencing and drug susceptibility testing is also likely to play a significant role in the determined level of resistance in each country. For example, in 2020 94% of new TB cases in the WHO Europe Region were tested for rifampicin resistance, compared to 50% in the African Region ( World Health Organization, 2022 ).
Emergence of resistance between hosts
Since the introduction of the first antituberculosis drug, streptomycin, in the 1950s, resistance to most new drugs has been identified as occurring within 5 to 10 years of their clinical use, with similar mutations occurring independently in different parts of the world (convergent evolution) ( Cohen et al., 2015 ; Manson et al., 2017 ; Brynildsrud et al., 2018 ). This phenomenon has occurred even more rapidly with recently introduced drugs.
An analysis of the world’s first comprehensively described extensively drug-resistant TB (XDR-TB, by historical definition of injectable and fluoroquinolone resistance) outbreak in Tugela Ferry, KwaZulu-Natal, South Africa, revealed that the drug resistance mutations carried by the strain had been acquired sequentially over 50 years. Genomic dating techniques revealed resistance developing broadly in the order in which drugs were introduced into clinical practice ( Cohen et al., 2015 ), and a similar pattern was demonstrated in a global collection of over 1500 lineage 4 strains ( Brynildsrud et al., 2018 ). This confirms the pattern established after the introduction of streptomycin in the mid-1940s, where clinical resistance was reported within two years ( Youmans et al., 1946 ) ( Figure 2 ). Two other studies of multiple global lineages have also shown the same order of resistance development, starting with isoniazid and streptomycin resistance, followed by rifampicin, fluoroquinolones and injectables ( Manson et al., 2017 ; Ektefaie et al., 2021 ). Interestingly, these studies did not show a correlation between the date of drug introduction and the date of resistance emerging. This is likely to represent the fact that other variables affect the development of resistance, such as the spontaneous rate at which mutations develop, in vivo fitness costs associated with resistance, and the clinical combinations in which drugs tended to be used (for example, while injectable drugs have been available since the 1950s they were much less commonly used than rifampicin and isoniazid).

Date of introduction to clinical use for antituberculous drugs (above line, denoted by solid black line) and estimated date of resistance emergence (below line, denoted by grey dashed line) ( Ektefaie et al., 2021 ).
Intriguingly, Rv0678 mutations likely to confer bedaquiline resistance have been identified long prior to the development of the drug van ( van Dorp et al., 2020 ). It has been hypothesised that this could have been selected for by the use of clofazimine, which was developed for the treatment of TB in the 1950s, although due to the development of more effective TB drugs was mostly used for the treatment of leprosy until it was repurposed for MDR-TB in the 2000s. The earliest emergence was dated to the beginning of the 18 th century, although interestingly this clade had an associated inactivating mutation in mmpL5 which was likely to counteract the resistant phenotype ( Sonnenkalb et al., 2021 ). However, later emergences in the late 19 th and early 20 th century still pre-date the use of any antituberculosis therapy, and may be due to other environmental stressors, including microbial antagonism in the environment before the transition to obligate pathogenicity. As one role of MmpL5 is efflux – especially of siderophores – this could include adapting to low iron availability or presence of a toxin for example. Overall, this demonstrates that, at least for bedaquiline and clofazimine, and potentially other new drugs, resistant bacteria may already exist in the environment and could be rapidly selected for as therapy is expanded.
Compensatory mutations
Many TB drugs target essential cellular processes, hence resistance-conferring mutations can come with a fitness cost, manifested as a slower growth rate in culture and reduced transmission within a population compared to wild-type strains. This has been best described for rpoB mutations conferring rifampicin resistance ( Gagneux et al., 2006 ; Knight et al., 2015 ). The most common isoniazid resistance mutation in katG (S315T) is thought to have only minimal fitness cost, while pyrazinamide resistance may have a fitness cost ( Pečerska et al., 2021 ). Most fluoroquinolone resistance-conferring mutations do not affect fitness, although impaired growth has been reported for the gyrA G88C and G88D mutations, although these occur infrequently in clinical isolates ( Emane et al., 2021 ).
The fitness cost imposed by rpoB mutations can be reversed by compensatory mutations, specifically mutations in rpoA and rpoC which encode two other subunits of the RNA polymerase enzyme (alpha and beta prime). These have been identified in in vitro culture experiments and additionally are seen at an increased prevalence in countries with a high burden of MDR-TB ( Comas et al., 2011 ; Merker et al., 2018 ; Trauner et al., 2021 ), linked to high rates of transmission of MDR-TB strains in some settings ( Gygli et al., 2021 ).
Emergence of resistance within hosts
It is likely that the ability of M. tuberculosis to rapidly acquire drug resistance while maintaining limited genomic variability over time is due the generation of low frequency within-host variation. This is not surprising given that M. tuberculosis infections typically last months to years and within-host bacterial populations may peak at over 10 9 colony forming units. Greater insight into this has been achieved through the adoption of high throughput sequencing, which has more recently enabled the identification significant within-host M. tuberculosis genetic diversity. Within-host diversity can in principle arise from mixed infection with multiple genetically distinct strains or within-host microevolution of a single infecting strain, or both ( Figure 3 ) ( Ford et al., 2012 ). At one extreme, up to 50 consensus-level SNP differences having been reported to occur over the duration of infection in patients with advanced disease when sampling from multiple body sites ( Lieberman et al., 2016 ). However in the majority of cases of M. tuberculosis infection, the genetic diversity is constrained by purifying selection that leads to loss of variants without a clear fitness advantage for this specialised pathogen which is adapted to a pathogenic lifestyle in the human host ( Figure 3 ).

Infection model showing how within-host genetic diversity may occur through mixed infection with genetically different strains or within-host evolution of a clonal infecting strain. Model of within-host evolution showing single dominant strain, with purifying selection leading to loss of variants with reduced fitness and multiple co-existing strains within lung.
Although most M. tuberculosis variants are lost over the course of infection, it is still possible for those that confer an advantage, such as drug resistance, to increase in prevalence and become the dominant clone over time. These resistant strains can then be transmitted leading to primary drug resistant infection in a new host. This may contrast with other bacteria where multiple variant strains may co-exist separately, as may be seen in non-specialised pathogens such as Pseudomonas aeruginosa ( Winstanley et al., 2016 ) or non-tuberculous mycobacteria ( Bryant et al., 2016 ; Shaw et al., 2019 ) in patients with cystic fibrosis, where there is an abnormal airway and immune environment ( Figure 3 ).
Understanding factors affecting overall within-host M. tuberculosis genetic diversity may offer insights into mechanisms controlling bacterial replication and evolution. Most studies to date rely on sequencing mycobacterial DNA extracted from culture to ensure sufficient DNA for sequencing, although this is likely to introduce bias by stochastic loss and selecting for bacterial subpopulations more suited to growth in culture ( Metcalfe et al., 2017 ). However, it has been demonstrated that culture-independent sequencing of M. tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture ( Nimmo et al., 2019 ; Shockey et al., 2019 ). As techniques for direct-from-sample sequencing improve, our understanding of within patient genetic diversity may therefore continue to develop.
From current work that had relied on sequencing from culture, one detailed study of five patients revealed that overall M. tuberculosis genomic diversity increased with disease severity and was particularly high in pre-mortem isolates from two patients, presumably due to high bacterial load ( O’Neill et al., 2015 ). The most sequence-diverse genes were those involved in production of cell envelope lipids. No evidence for a decrease in diversity during treatment or any effect of M. tuberculosis lineage or drug resistance profile was found, while HIV statuses were not available for analysis. An analysis of 200 patients from eight publicly available studies reporting patients who failed treatment found that genes associated with antibiotic resistance displayed highest diversity, while the within-host diversity across remaining gene classes ( in vitro essential, non-essential, PE/PPE genes and antigen genes) seemed unaffected ( Vargas et al., 2021 ). South African cohort studies revealed greater genetic diversity in patients with cavitary disease, infection with lineage 2 strains and absence of second-line drug resistance, although no association between time to positivity in culture and diversity ( Nimmo et al., 2020 ). This suggests that diversity may be more influenced by higher intrinsic mutation rates (as seen with lineage 2), variable drug penetration (in cavitary disease) or impaired immune control (in untreated HIV) than bacterial population size. However, there was no association between diversity and clinical outcomes at six months.
Mixed populations of wild-type alleles and resistance-associated variants (RAVs) confer heteroresistance, where populations of resistant and susceptible bacteria co-exist within the same host. This may occur as the result of differential drug penetration to spatially and pathologically distinct lung regions ( Dheda et al., 2018 ) leading in effect to monotherapy and subsequent resistance acquisition or survival of susceptible bacteria. Baseline genetic heteroresistance appears to be particularly common for bedaquiline (up to 60%) ( Nimmo et al., 2020 ) and fluoroquinolones (11-26%) ( Operario et al., 2017 ; Nimmo et al., 2020 ).
Several case reports have identified heterozygous RAVs that have increased in frequency over the course of treatment ( Sun et al., 2012 ; Eldholm et al., 2014 ; Trauner et al., 2017 ) leading to fixed resistance, including variants originally identified at <1% frequency ( Vos et al., 2019 ). A retrospective deep sequencing study identified very low frequency RAVs (<1%) predating acquired phenotypic resistance ( Engelthaler et al., 2019 ). However, due to high levels of turnover of low-frequency variants, it may be difficult to predict which heterozygous RAVs are likely to persist or become fixed and which ones will disappear. In a prospective cohort study of almost 400 patients, most with heterozygous RAVs detectable on WGS with sequential isolates available and sensitivity to detect variants above 5% frequency demonstrated RAV persistence or fixation (17/20, 85%) ( Nimmo et al., 2020 ). However, only one case of a very low frequency RAV (<5%) expanding to cause resistance was identified. Another study showed no effect on treatment outcome amongst patients with RAVs at <1% frequency ( Chen et al., 2021 ), while modelling from multiple cohort studies suggests that variants at ≥19% frequency predicted subsequent fixation ( Vargas et al., 2021 ).
Taken together, the current evidence suggests that the significance of heterozygous RAVs is likely to depend on their frequency, with much greater clinical significance of those at higher frequency (>15-20%) than lower frequency (especially <5%). While heterozygous RAVs are likely to be variably identified by current diagnostics ( Ng et al., 2019 ; Rigouts et al., 2019 ) with newer sequencing-based techniques offering good sensitivity even for RAVs identified at very low frequency, establishing how to interpret low frequency heterozygous RAVs is going to become an important clinical decision.
Implications for diagnostics
As many countries move towards genomic-based methods such as molecular PCR-based tools (for instance Xpert MTB/RIF), and progressively to targeted sequencing and whole genome sequencing (WGS) for diagnosis of M. tuberculosis infection and identification of drug resistance, it is important to be aware of the implications of the evolution of resistance. Currently, understanding of resistance-conferring mutations is inevitably limited, particularly for newer drugs, despite recent large global studies ( The CRyPTIC Consortium, 2022 ). The clearest example of a targeted molecular tool creating its own selective pressure has been demonstrated in Eswatini where the non-canonical rifampicin resistance-conferring mutation, rpoB I491F, which falls outside the rifampicin-resistance determining region became dominant ( Figure 4A ) ( Sanchez-Padilla et al., 2015 ).

Schematic diagrams of (A) rpoB gene codons 426-504 showing rifampicin resistance determining region, coverage of Xpert MTB/RIF probes (modified from ( Chakravorty et al., 2017 ) under CC BY 4.0 licence) and location of I491 codon, and (B) Rv0678 gene showing distribution of reported gene substitutions, coloured by association with resistance (reproduced from ( Ismail et al., 2021 ) under CC BY 4.0 licence).
This was first identified following the Eswatini 2009 drug resistance survey, which revealed a surprisingly high rate of MDR-TB (7.7% in previously untreated patients and 33.8% in previously treated patients) ( Sanchez-Padilla et al., 2012 ). Xpert MTB/RIF was implemented in 2012 to enable rapid diagnosis of MDR-TB ( Sikhondze et al., 2015 ), but in 2015 a detailed genetic and phenotypic analysis of strains stored from the 2009 survey showed that 30% of rifampicin resistance was actually conferred by a the rpoB I491F mutation, which is not identified by Xpert MTB/RIF and such strains would therefore be reported as rifampicin susceptible ( Sanchez-Padilla et al., 2015 ). Patients infected with such strains would therefore be treated with an ineffective standard drug-susceptible regimen for their rifampicin-resistant infection. By the time of the next drug resistance survey in 2017, 56% of rifampicin resistance was conferred by the I491F mutation ( World Health Organization, 2020 ). The Eswatini National Tuberculosis Control Programme has since proposed presumptively treating all isoniazid-resistant TB as MDR-TB until phenotypic testing has been completed, as most I491F mutations are present in isoniazid resistant strains ( Ardizzoni et al., 2021 ).
Finally, resistance to new drugs such as bedaquiline and delamanid is caused by many individually rare mutations that do not provide statistical power for genotype-phenotype correlation in the way that has been performed for most first-line drugs. For example, in the Rv0678 gene responsible for most clinical bedaquiline resistance, mutations are spread throughout the gene with no clear resistance-conferring hotspot ( Figure 4B ). Additionally, there is not a clear separation of minimum inhibitory concentrations of bedaquiline between wild type and resistant isolates, which is likely to complicate attempts to categorise individual mutations as susceptible or resistant, with many likely to fall near the critical concentration and be vulnerable to technical variation. A pragmatic approach may be to use molecular or genetic methods to screen resistance-associated genes for variants, which are rare amongst susceptible isolates, followed by phenotypic evaluation of isolates containing mutants.
Conclusions
M. tuberculosis has shown a remarkable ability to develop resistance to all antituberculosis drugs that have been developed, including those brought into clinical use for DR-TB in recent years such as bedaquiline, linezolid and delamanid/pretomanid. It is important to bear in mind these drugs remain highly effective in the vast majority of patients with DR-TB and have undoubtedly been responsible for the major improvements in DR-TB outcomes that have been seen in the last 10 years ( World Health Organization, 2022 ). However, examples such as the spread of XDR-TB across South Africa, first identified as the Tugela Ferry outbreak, and the rapid amplification of the rpoB I491F mutation amongst M. tuberculosis strains in Eswatini, highlight that this progress cannot be taken for granted ( Gandhi et al., 2006 ; Sanchez-Padilla et al., 2015 ). Progression towards the World Health Organization’s End TB targets of a 90% reduction in TB transmission between 2015 and 2035 will require strict control of the spread of DR-TB, which require highly effective drugs to be available rapidly to those infected.
To achieve this, important strategic decisions will be required. While there are now a number of exciting new drug candidates progressing through the TB drug development pipeline, their impact will remain uncertain. The effectiveness of currently available drugs depends on limiting the spread of resistance to them. It is therefore questionable whether effective drugs for DR-TB such as bedaquiline and pretomanid should be incorporated into drug-susceptible TB (DS-TB) regimens, which may increase the spread of resistance and reduce their effectiveness for DR-TB, unless their overall benefits to patients with TB and progress towards elimination is outweighed by improvements in DS-TB treatment. This needs to be accounted for when evaluating the results of trials such as SimpliciTB (ClinicalTrials.gov Identifier: {"type":"clinical-trial","attrs":{"text":"NCT03338621","term_id":"NCT03338621"}} NCT03338621 ), where bedaquiline and pretomanid are used to reduce DS-TB treatment duration from six to four months, which was already been demonstrated to be possible using rifapentine and moxifloxacin ( Dorman et al., 2021 ), or adopting a stratified treatment approach for some patients ( Imperial et al., 2018 ).
In addition, it is essential not to assume susceptibility to drugs and TB programmes should aim to perform susceptibility testing for all drugs that are included in treatment regimens. It is now clearly demonstrated that there is a pre-existing pool of bedaquiline resistance and it can therefore be expected to occur in patients without any clear risk factors for resistance ( Beckert et al., 2020 ; van Dorp et al., 2020 ; Ismail et al., 2022 ) Genotypic susceptibility testing is clearly very effective in many cases, but it is important to be aware of the limitations. While the limited genotypic-phenotypic understanding for new drugs such as bedaquiline and delamanid/pretomanid will be one challenge, the spread of rpoB I491F shows how significant the selective pressure from diagnostics can be.
The most effective strategy is therefore going to require greater understanding of how resistance develops within patients with a view to preventing its occurrence. This will need to be backed up by preventing spread of resistance through use of up-front susceptibility testing for all drugs – using combined genotypic and phenotypic methods – along with ongoing surveillance for development of resistance and changes to the prevalence of resistance-conferring mutations, and strategic use of available medications to maximise benefit to individual patients as well as the End TB strategy.
Author contributions
Conceptualisation: CN;Writing – original draft: CN; Writing – review and editing: CN, JaM, VF, JoM, JE, HP, and EJ; Visualisation: CN, VF, JoM, and HP; Supervision: EOJ; All authors contributed to the article and approved the submitted version.
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (CC2169), the UK Medical Research Council (CC2169), and the Wellcome Trust (CC2169).
Acknowledgments
Figures 2 , 3 and 4 created with BioRender.com . This research was funded in part by the Wellcome Trust (CC2169). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The authors would like to thank Joanna Evans (Systems Chemical Biology of Infection and Resistance Laboratory, Francis Crick Institute) for critically reading the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
- Ardizzoni E., Ariza E., Mulengwa D., Mpala Q., Tour R., de L., et al.. (2021). Thin-Layer-Agar-Based direct phenotypic drug susceptibility testing on sputum in eswatini rapidly detects mycobacterium tuberculosis growth and rifampicin resistance otherwise missed by WHO-endorsed diagnostic tests . Antimicrob. Agents Ch 65 , e02263–e02220. doi: 10.1128/aac.02263-20 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Asante-Poku A., Yeboah-Manu D., Otchere I. D., Aboagye S. Y., Stucki D., Hattendorf J., et al.. (2015). Mycobacterium africanum is associated with patient ethnicity in Ghana . PloS Negl. Trop. D 9 , e3370. doi: 10.1371/journal.pntd.0003370 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K. S., Wilson T., et al.. (1994). inhA, a gene encoding a target for isoniazid and ethionamide in mycobacterium tuberculosis . Science 263 , 227–230. doi: 10.1126/science.8284673 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Beckert P., Sanchez-Padilla E., Merker M., Dreyer V., Kohl T. A., Utpatel C., et al.. (2020). MDR m. Tuberculosis outbreak clone in eswatini missed by xpert has elevated bedaquiline resistance dated to the pre-treatment era . Genome Med. 12 , 104. doi: 10.1186/s13073-020-00793-8 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bergval I., Kwok B., Schuitema A., Kremer K., van Soolingen D., Klatser P., et al.. (2012). Pre-existing isoniazid resistance, but not the genotype of mycobacterium tuberculosis drives rifampicin resistance codon preference in vitro . PloS One 7 , e29108. doi: 10.1371/journal.pone.0029108 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bergval I. L., Schuitema A. R. J., Klatser P. R., Anthony R. M. (2009). Resistant mutants of mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance . J. Antimicrob. Chemoth 64 , 515–523. doi: 10.1093/jac/dkp237 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Billington O. J., McHugh T. D., Gillespie S. H. (1999). Physiological cost of rifampin resistance induced In vitro in mycobacterium tuberculosis . Antimicrob. Agents Ch 43 , 1866–1869. doi: 10.1128/aac.43.8.1866 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Boritsch E. C., Khanna V., Pawlik A., Honoré N., Navas V. H., Ma L., et al.. (2016). Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria . Proc. Natl. Acad. Sci. 113 , 9876–9881. doi: 10.1073/pnas.1604921113 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bos K. I., Harkins K. M., Herbig A., Coscolla M., Weber N., Comas I., et al.. (2014). Pre-Columbian mycobacterial genomes reveal seals as a source of new world human tuberculosis . Nature 514 , 494–497. doi: 10.1038/nature13591 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bryant J. M., Grogono D. M., Rodriguez-Rincon D., Everall I., Brown K. P., Moreno P., et al.. (2016). Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium . Science 354 , 751–757. doi: 10.1126/science.aaf8156 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Brynildsrud O. B., Pepperell C. S., Suffys P., Grandjean L., Monteserin J., Debech N., et al.. (2018). Global expansion of mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation . Sci. Adv. 4 , eaat5869. doi: 10.1126/sciadv.aat5869 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chakravorty S., Simmons A. M., Rowneki M., Parmar H., Cao Y., Ryan J., et al.. (2017). The new xpert MTB/RIF ultra: Improving detection of mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-Care testing . Mbio 8 , e00812–e00817. doi: 10.1128/mbio.00812-17 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen Y., Jiang Q., Zou J., Yang T., Liu Q., Luo G., et al.. (2021). Deep whole-genome sequencing reveals no evidence for heteroresistance influencing treatment outcomes among drug-susceptible tuberculosis patients . Tuberculosis 130 , 102120. doi: 10.1016/j.tube.2021.102120 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chiner-Oms Á., Sánchez-Busó L., Corander J., Gagneux S., Harris S. R., Young D., et al.. (2019). Genomic determinants of speciation and spread of the mycobacterium tuberculosis complex . Sci. Adv. 5 , eaaw3307. doi: 10.1126/sciadv.aaw3307 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cohen K. A., Abeel T., McGuire A. M., Desjardins C. A., Munsamy V., Shea T. P., et al.. (2015). Evolution of extensively drug-resistant tuberculosis over four decades: Whole genome sequencing and dating analysis of mycobacterium tuberculosis isolates from KwaZulu-natal . PloS Med. 12 , e1001880. doi: 10.1371/journal.pmed.1001880 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al.. (1998). Deciphering the biology of mycobacterium tuberculosis from the complete genome sequence . Nature 393 , 537–544. doi: 10.1038/31159 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Comas I., Borrell S., Roetzer A., Rose G., Malla B., Kato-Maeda M., et al.. (2011). Whole-genome sequencing of rifampicin-resistant m. tuberculosis strains identifies compensatory mutations in RNA polymerase . Nat. Genet. 44 , 106–110. doi: 10.1038/ng.1038 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Comas I., Coscolla M., Luo T., Borrell S., Holt K. E., Kato-Maeda M., et al.. (2013). Out-of-Africa migration and neolithic co-expansion of mycobacterium tuberculosis with modern humans . Nat. Genet. 45 , 1176–1182. doi: 10.1038/ng.2744 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- David H. L. (1970). Probability distribution of drug-resistant mutants in unselected populations of mycobacterium tuberculosis . Appl. Microbiol. 20 , 810–814. doi: 10.1128/am.20.5.810-814.1970 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dheda K., Lenders L., Magombedze G., Srivastava S., Raj P., Arning E., et al.. (2018). Drug penetration gradients associated with acquired drug resistance in tuberculosis patients . Am. J. Resp. Crit. Care 198 , 1208–1219. doi: 10.1164/rccm.201711-2333oc [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dookie N., Rambaran S., Padayatchi N., Mahomed S., Naidoo K. (2018). Evolution of drug resistance in mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care . J. Antimicrob. Chemoth 73 , 1138–1151. doi: 10.1093/jac/dkx506 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dorman S. E., Nahid P., Kurbatova E. V., Phillips P. P. J., Bryant K., Dooley K. E., et al.. (2021). Four-month rifapentine regimens with or without moxifloxacin for tuberculosis . New Engl. J. Med. 384 , 1705–1718. doi: 10.1056/nejmoa2033400 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- van Dorp L., Nimmo C., Ortiz A. T., Pang J., Acman M., Tan C. C. S., et al.. (2020) Detection of a bedaquiline / clofazimine resistance reservoir in mycobacterium tuberculosis predating the antibiotic era . Biorxiv . doi: 10.1101/2020.10.06.328799 [ CrossRef ] [ Google Scholar ]
- Ektefaie Y., Dixit A., Freschi L., Farhat M. R. (2021). Globally diverse mycobacterium tuberculosis resistance acquisition: A retrospective geographical and temporal analysis of whole genome sequences . Lancet Microbe 2 , e96–e104. doi: 10.1016/s2666-5247(20)30195-6 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Eldholm V., Balloux F. (2016). Antimicrobial resistance in mycobacterium tuberculosis: The odd one out . Trends Microbiol. 24 , 637–648. doi: 10.1016/j.tim.2016.03.007 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Eldholm V., Norheim G., Lippe B.v. d., Kinander W., Dahle U. R., Caugant D. A., et al.. (2014). Evolution of extensively drug-resistant mycobacterium tuberculosis from a susceptible ancestor in a single patient . Genome Biol. 15 , 490. doi: 10.1186/s13059-014-0490-3 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Eldholm V., Pettersson J. H.-O., Brynildsrud O. B., Kitchen A., Rasmussen E. M., Lillebaek T., et al.. (2016). Armed conflict and population displacement as drivers of the evolution and dispersal of mycobacterium tuberculosis . Proc. Natl. Acad. Sci. 113 , 13881–13886. doi: 10.1073/pnas.1611283113 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Emane A. K. A., Guo X., Takiff H. E., Liu S. (2021). Drug resistance, fitness and compensatory mutations in mycobacterium tuberculosis . Tuberculosis 129 , 102091. doi: 10.1016/j.tube.2021.102091 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Engelthaler D. M., Streicher E. M., Kelley E. J., Allender C. J., Wiggins K., Jimenez D., et al.. (2019). Minority mycobacterium tuberculosis genotypic populations as an indicator of subsequent phenotypic resistance . Am. J. Resp. Cell Mol. 61 , 789–791. doi: 10.1165/rcmb.2019-0178le [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fishbein S., Wyk N., Warren R. M., Sampson S. L. (2015). Phylogeny to function: PE/PPE protein evolution and impact on mycobacterium tuberculosis pathogenicity . Mol. Microbiol. 96 , 901–916. doi: 10.1111/mmi.12981 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ford C. B., Lin P. L., Chase M. R., Shah R. R., Iartchouk O., Galagan J., et al.. (2011). Use of whole genome sequencing to estimate the mutation rate of mycobacterium tuberculosis during latent infection . Nat. Genet. 43 , 482–486. doi: 10.1038/ng.811 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ford C. B., Shah R. R., Maeda M. K., Gagneux S., Murray M. B., Cohen T., et al.. (2013). Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis . Nat. Genet. 45 , 784–790. doi: 10.1038/ng.2656 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ford C., Yusim K., Ioerger T., Feng S., Chase M., Greene M., et al.. (2012). Mycobacterium tuberculosis – heterogeneity revealed through whole genome sequencing . Tuberculosis 92 , 194–201. doi: 10.1016/j.tube.2011.11.003 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gagneux S. (2018). Ecology and evolution of mycobacterium tuberculosis . Nat. Rev. Microbiol. 16 , 202–213. doi: 10.1038/nrmicro.2018.8 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gagneux S., Long C. D., Small P. M., Van T., Schoolnik G. K., Bohannan B. J. M. (2006). The competitive cost of antibiotic resistance in mycobacterium tuberculosis . Science 312 , 1944–1946. doi: 10.1126/science.1124410 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gandhi N. R., Moll A., Sturm A. W., Pawinski R., Govender T., Lalloo U., et al.. (2006). Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of south Africa . Lancet 368 , 1575–1580. doi: 10.1016/s0140-6736(06)69573-1 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gomez J. E., Kaufmann-Malaga B. B., Wivagg C. N., Kim P. B., Silvis M. R., Renedo N., et al.. (2017). Ribosomal mutations promote the evolution of antibiotic resistance in a multidrug environment . Elife 6 , e20420. doi: 10.7554/elife.20420 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Grandjean L., Iwamoto T., Lithgow A., Gilman R. H., Arikawa K., Nakanishi N., et al.. (2015). The association between mycobacterium tuberculosis genotype and drug resistance in Peru . PloS One 10 , e0126271. doi: 10.1371/journal.pone.0126271 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Guerra-Assunção J., Crampin A., Houben R., Mzembe T., Mallard K., Coll F., et al.. (2015). Large-Scale whole genome sequencing of m. tuberculosis provides insights into transmission in a high prevalence area . Elife 4 , e05166. doi: 10.7554/elife.05166 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gupta R. S., Lo B., Son J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus mycobacterium into an emended genus mycobacterium and four novel genera . Front. Microbiol. 9 . doi: 10.3389/fmicb.2018.00067 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gygli S. M., Loiseau C., Jugheli L., Adamia N., Trauner A., Reinhard M., et al.. (2021). Prisons as ecological drivers of fitness-compensated multidrug-resistant mycobacterium tuberculosis . Nat. Med. 27 , 1171–1177. doi: 10.1038/s41591-021-01358-x [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hakamata M., Takihara H., Iwamoto T., Tamaru A., Hashimoto A., Tanaka T., et al.. (2020). Higher genome mutation rates of Beijing lineage of mycobacterium tuberculosis during human infection . Sci. Rep-uk 10 , 17997. doi: 10.1038/s41598-020-75028-2 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hartkoorn R. C., Uplekar S., Cole S. T. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in mycobacterium tuberculosis . Antimicrob. Agents Ch 58 , 2979–2981. doi: 10.1128/aac.00037-14 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Haver H. L., Chua A., Ghode P., Lakshminarayana S. B., Singhal A., Mathema B., et al.. (2015). Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous In vitro -selected PA-824-Resistant mutants of mycobacterium tuberculosis . Antimicrob. Agents Ch 59 , 5316–5323. doi: 10.1128/aac.00308-15 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hershkovitz I., Donoghue H. D., Minnikin D. E., Besra G. S., Lee O. Y.-C., Gernaey A. M., et al.. (2008). Detection and molecular characterization of 9000-Year-Old mycobacterium tuberculosis from a neolithic settlement in the Eastern Mediterranean . PloS One 3 , e3426. doi: 10.1371/journal.pone.0003426 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Huitric E., Verhasselt P., Koul A., Andries K., Hoffner S., Andersson D. I. (2010). Rates and mechanisms of resistance development in mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor . Antimicrob. Agents Ch 54 , 1022–1028. doi: 10.1128/aac.01611-09 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Imperial M. Z., Nahid P., Phillips P. P. J., Davies G. R., Fielding K., Hanna D., et al.. (2018). A patient-level pooled analysis of treatment-shortening regimens for drug-susceptible pulmonary tuberculosis . Nat. Med. 24 , 1708–1715. doi: 10.1038/s41591-018-0224-2 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ismail N. A., Omar S. V., Moultrie H., Bhyat Z., Conradie F., Enwerem M., et al.. (2022). Assessment of epidemiological and genetic characteristics and clinical outcomes of resistance to bedaquiline in patients treated for rifampicin-resistant tuberculosis: a cross-sectional and longitudinal study . Lancet Infect. Dis. 22 , 496–506. doi: 10.1016/s1473-3099(21)00470-9 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ismail N., Rivière E., Limberis J., Huo S., Metcalfe J. Z., Warren R. M., et al.. (2021). Genetic variants and their association with phenotypic resistance to bedaquiline in mycobacterium tuberculosis: a systematic review and individual isolate data analysis . Lancet Microbe 2 , e604–e616. doi: 10.1016/s2666-5247(21)00175-0 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Johnson E. O., Office E., Kawate T., Orzechowski M., Hung D. T. (2020). Large-Scale chemical-genetic strategy enables the design of antimicrobial combination chemotherapy in mycobacteria . ACS Infect. Dis. 6 , 56–63. doi: 10.1021/acsinfecdis.9b00373 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kana B. D., Abrahams G. L., Sung N., Warner D. F., Gordhan B. G., Machowski E. E., et al.. (2010). Role of the DinB homologs Rv1537 and Rv3056 in mycobacterium tuberculosis . J. Bacteriol 192 , 2220–2227. doi: 10.1128/jb.01135-09 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kay G. L., Sergeant M. J., Zhou Z., Chan J. Z.-M., Millard A., Quick J., et al.. (2015). Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe . Nat. Commun. 6 , 6717. doi: 10.1038/ncomms7717 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Knight G. M., Colijn C., Shrestha S., Fofana M., Cobelens F., White R. G., et al.. (2015). The distribution of fitness costs of resistance-conferring mutations is a key determinant for the future burden of drug-resistant tuberculosis: A model-based analysis . Clin. Infect. Dis. 61 , S147–S154. doi: 10.1093/cid/civ579 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lieberman T. D., Wilson D., Misra R., Xiong L. L., Moodley P., Cohen T., et al.. (2016). Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated mycobacterium tuberculosis . Nat. Med. 22 , 1470–1474. doi: 10.1038/nm.4205 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Manson A. L., Cohen K. A., Abeel T., Desjardins C. A., Armstrong D. T., Barry C. E., et al.. (2017). Genomic analysis of globally diverse mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance . Nat. Genet. 49 , 395–402. doi: 10.1038/ng.3767 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- McGrath M., van Pittius N. C. G., van Helden P. D., Warren R. M., Warner D. F. (2014). Mutation rate and the emergence of drug resistance in mycobacterium tuberculosis . J. Antimicrob. Chemoth 69 , 292–302. doi: 10.1093/jac/dkt364 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Merker M., Barbier M., Cox H., Rasigade J.-P., Feuerriegel S., Kohl T. A., et al.. (2018). Compensatory evolution drives multidrug-resistant tuberculosis in central Asia . Elife 7 , e38200. doi: 10.7554/elife.38200 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Metcalfe J. Z., Streicher E., Theron G., Colman R. E., Penaloza R., Allender C., et al.. (2017). Mycobacterium tuberculosis subculture results in loss of potentially clinically relevant heteroresistance . Antimicrob. Agents Ch 61 ( 11 ):e00888–17. doi: 10.1128/aac.00888-17 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Morlock G. P., Metchock B., Sikes D., Crawford J. T., Cooksey R. C. (2003). ethA , inhA , and katG loci of ethionamide-resistant clinical mycobacterium tuberculosis isolates . Antimicrob. Agents Ch 47 , 3799–3805. doi: 10.1128/aac.47.12.3799-3805.2003 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ng K. C. S., Supply P., Cobelens F. G. J., Gaudin C., Gonzalez-Martin J., de Jong B. C., et al.. (2019). How well do routine molecular diagnostics detect rifampin heteroresistance in mycobacterium tuberculosis ? J. Clin. Microbiol. 57 , e00717–e00719. doi: 10.1128/jcm.00717-19 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nimmo C., Brien K., Millard J., Grant A. D., Padayatchi N., Pym A. S., et al.. (2020). Dynamics of within-host mycobacterium tuberculosis diversity and heteroresistance during treatment . Ebiomedicine 55 , 102747. doi: 10.1016/j.ebiom.2020.102747 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nimmo C., Shaw L. P., Doyle R., Williams R., Brien K., Burgess C., et al.. (2019). Whole genome sequencing mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture . BMC Genomics 20 , 389. doi: 10.1186/s12864-019-5782-2 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- O’Neill M. B., Mortimer T. D., Pepperell C. S. (2015). Diversity of mycobacterium tuberculosis across evolutionary scales . PloS Pathog. 11 , e1005257. doi: 10.1371/journal.ppat.1005257 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- O’Neill M. B., Shockey A., Zarley A., Aylward W., Eldholm V., Kitchen A., et al.. (2019). Lineage specific histories of mycobacterium tuberculosis dispersal in Africa and Eurasia . Mol. Ecol. 28 , 3241–3256. doi: 10.1111/mec.15120 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Operario D. J., Koeppel A. F., Turner S. D., Bao Y., Pholwat S., Banu S., et al.. (2017). Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis . PloS One 12 , e0176522. doi: 10.1371/journal.pone.0176522 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ortiz A. T., Coronel J., Vidal J. R., Bonilla C., Moore D. A. J., Gilman R. H., et al.. (2021). Genomic signatures of pre-resistance in mycobacterium tuberculosis . Nat. Commun. 12 , 7312. doi: 10.1038/s41467-021-27616-7 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- O’Sullivan D. M., McHugh T. D., Gillespie S. H. (2008). The effect of oxidative stress on the mutation rate of mycobacterium tuberculosis with impaired catalase/peroxidase function . J. Antimicrob. Chemoth 62 , 709–712. doi: 10.1093/jac/dkn259 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Parwati I., van Crevel R., van Soolingen D. (2010). Possible underlying mechanisms for successful emergence of the mycobacterium tuberculosis Beijing genotype strains . Lancet Infect. Dis. 10 , 103–111. doi: 10.1016/s1473-3099(09)70330-5 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Pečerska J., Kühnert D., Meehan C. J., Coscollá M., Jong B. C., Gagneux S., et al.. (2021). Quantifying transmission fitness costs of multi-drug resistant tuberculosis . Epidemics-neth 36 , 100471. doi: 10.1016/j.epidem.2021.100471 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Pym A. S., Saint-Joanis B., Cole S. T. (2002). Effect of katG mutations on the virulence of mycobacterium tuberculosis and the implication for transmission in humans . Infect. Immun. 70 , 4955–4960. doi: 10.1128/iai.70.9.4955-4960.2002 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Rigouts L., Miotto P., Schats M., Lempens P., Cabibbe A. M., Galbiati S., et al.. (2019). Fluoroquinolone heteroresistance in mycobacterium tuberculosis: detection by genotypic and phenotypic assays in experimentally mixed populations . Sci. Rep-uk 9 , 11760. doi: 10.1038/s41598-019-48289-9 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sanchez-Padilla E., Dlamini T., Ascorra A., Rüsch-Gerdes S., Tefera Z. D., Calain P., et al.. (2012). High prevalence of multidrug-resistant tuberculosis, swazilan –2010 . Emerg. Infect. Dis. 18 , 29–37. doi: 10.3201/eid1801.110850 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sanchez-Padilla E., Merker M., Beckert P., Jochims F., Dlamini T., Kahn P., et al.. (2015). Detection of drug-resistant tuberculosis by xpert MTB/RIF in Swaziland . New Engl. J. Med. 372 , 1181–1182. doi: 10.1056/nejmc1413930 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Scorpio A., Zhang Y. (1996). Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus . Nat. Med. 2 , 662–667. doi: 10.1038/nm0696-662 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Shaw L. P., Doyle R. M., Kavaliunaite E., Spencer H., Balloux F., Dixon G., et al.. (2019). Children with cystic fibrosis are infected with multiple subpopulations of mycobacterium abscessus with different antimicrobial resistance profiles . Clin. Infect. Dis. Off. Publ Infect. Dis. Soc. Am. 69 , 1678–1686. doi: 10.1093/cid/ciz069 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Shockey A. C., Dabney J., Pepperell C. S. (2019). Effects of host, sample, and in vitro culture on genomic diversity of pathogenic mycobacteria . Front. Genet. 10 . doi: 10.3389/fgene.2019.00477 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sikhondze W., Dlamini T., Khumalo D., Maphalala G., Dlamini S., Zikalala T., et al.. (2015). Countrywide roll-out of xpert® MTB/RIF in Swaziland: the first three years of implementation . Public Heal Action 5 , 140–146. doi: 10.5588/pha.15.0001 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Silva P. E. A. D., Palomino J. C. (2011). Molecular basis and mechanisms of drug resistance in mycobacterium tuberculosis: classical and new drugs . J. Antimicrob. Chemoth 66 , 1417–1430. doi: 10.1093/jac/dkr173 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sonnenkalb L., Carter J., Spitaleri A., Iqbal Z., Hunt M., Malone K., et al.. (2021). Deciphering bedaquiline and clofazimine resistance in tuberculosis: An evolutionary medicine approach . Biorxiv . doi: 10.1101/2021.03.19.436148 Available at: https://www.biorxiv.org/content/10.1101/2021.03.19.436148v4 [ CrossRef ] [ Google Scholar ]
- Soolingen D. V., Hoogenboezem T., Haas P. E. W. D., Hermans P. W. M., Koedam M. A., Teppema K. S., et al.. (1997). A novel pathogenic taxon of the mycobacterium tuberculosis complex, canetti: Characterization of an exceptional isolate from Africa . Int. J. Syst. Evol. Micr 47 , 1236–1245. doi: 10.1099/00207713-47-4-1236 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Stoffels K., Mathys V., Fauville-Dufaux M., Wintjens R., Bifani P. (2012). Systematic analysis of pyrazinamide-resistant spontaneous mutants and clinical isolates of mycobacterium tuberculosis . Antimicrob. Agents Ch 56 , 5186–5193. doi: 10.1128/aac.05385-11 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sun G., Luo T., Yang C., Dong X., Li J., Zhu Y., et al.. (2012). Dynamic population changes in mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients . J. Infect. Dis. 206 , 1724–1733. doi: 10.1093/infdis/jis601 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Suzuki Y., Katsukawa C., Tamaru A., Abe C., Makino M., Mizuguchi Y., et al.. (1998). Detection of kanamycin-resistant mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene . J. Clin. Microbiol. 36 , 1220–1225. doi: 10.1128/jcm.36.5.1220-1225.1998 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Takiff H. E., Salazar L., Guerrero C., Philipp W., Huang W. M., Kreiswirth B., et al.. (1994). Cloning and nucleotide sequence of mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations . Antimicrob. Agents Ch 38 , 773–780. doi: 10.1128/aac.38.4.773 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Telenti A., Imboden P., Marchesi F., Matter L., Schopfer K., Bodmer T., et al.. (1993). Detection of rifampicin-resistance mutations in mycobacterium tuberculosis . Lancet 341 , 647–651. doi: 10.1016/0140-6736(93)90417-f [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Telenti A., Philipp W. J., Sreevatsan S., Bernasconi C., Stockbauer K. E., Wieles B., et al.. (1997). The emb operon, a gene cluster of mycobacterium tuberculosis involved in resistance to ethambutol . Nat. Med. 3 , 567–570. doi: 10.1038/nm0597-567 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- The CRyPTIC Consortium (2022). A data compendium associating the genomes of 12,289 mycobacterium tuberculosis isolates with quantitative resistance phenotypes to 13 antibiotics . PloS Biol. 20 , e3001721. doi: 10.1371/journal.pbio.3001721 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tortoli E., Brown-Elliott B. A., Chalmers J. D., Cirillo D. M., Daley C. L., Emler S., et al.. (2019). Same meat, different gravy: ignore the new names of mycobacteria . Eur. Respir. J. 54 , 1900795. doi: 10.1183/13993003.00795-2019 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Trauner A., Banaei-Esfahani A., Gygli S. M., Warmer P., Feldmann J., Zampieri M., et al.. (2021). Expression dysregulation as a mediator of fitness costs in antibiotic resistance . Antimicrob. Agents Ch 65 , e00504–e00521. doi: 10.1128/aac.00504-21 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Trauner A., Liu Q., Via L. E., Liu X., Ruan X., Liang L., et al.. (2017). The within-host population dynamics of mycobacterium tuberculosis vary with treatment efficacy . Genome Biol. 18 , 71. doi: 10.1186/s13059-017-1196-0 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Vargas R., Freschi L., Marin M., Epperson L. E., Smith M., Oussenko I., et al.. (2021). In-host population dynamics of mycobacterium tuberculosis complex during active disease . Elife 10 , e61805. doi: 10.7554/elife.61805 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Vos M., Ley S. D., Wiggins K. B., Derendinger B., Dippenaar A., Grobbelaar M., et al.. (2019). Bedaquiline micro-heteroresistance after tuberculosis treatment cessation . New Engl. J. Med. 380 , 2178–2180. doi: 10.1056/nejmc1815121 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Werngren J., Hoffner S. E. (2003). Drug-susceptible mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate . J. Clin. Microbiol. 41 , 1520–1524. doi: 10.1128/jcm.41.4.1520-1524.2003 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Winstanley C., O’Brien S., Brockhurst M. A. (2016). Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections . Trends Microbiol. 24 , 327–337. doi: 10.1016/j.tim.2016.01.008 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- World Health Organization (2020) Global tuberculosis report 2019 . Available at: https://www.who.int/publications/i/item/9789241565714 (Accessed April 27, 2022).
- World Health Organization (2022) Global tuberculosis programme: tuberculosis data . Available at: https://www.who.int/teams/global-tuberculosis-programme/data (Accessed April 27, 2022).
- Youmans G. P., Williston E. H., Feldman W. H., Hinshaw H. C. (1946). Increase in resistance of tubercle bacilli to streptomycin; a preliminary report . Proc. Staff Meet Mayo Clin. 21 , 126. [ PubMed ] [ Google Scholar ]
Universities in Omsk, Russia - Rankings & Reviews -
- 06 Mar, 2024: Scimago Institutions Rankings updated with Omsk State Technical University ranked highest among 3 listed universities in Omsk.
- 19 Oct, 2023: THE Times Higher Education, UK published most recent results of THE World University Rankings by Subject (Business and Economics) . Omsk State Technical University achieves position 1001.
- 27 Sep, 2023: Latest THE World University Rankings from THE Times Higher Education, UK. Omsk State Technical University in position #1501.
- 31 Jul, 2023: Latest Webometrics Ranking Web of Universities from Webometrics. 6 universities from Omsk appear in this ranking.
Rankings of universities in Omsk, Russia 2024

Omsk State Technical University
- University rankings - Omsk State Technical University
| THE World University Rankings | |||
| Scimago Institutions Rankings | |||
| THE Emerging Economies University Ranking - Times Higher Education | |||
| Show 6 more rankings of Omsk State Technical University and subject specific rankings for 1 subjects | |||

Omsk State Medical Academy

- University rankings - Omsk State Medical Academy
| Scimago Institutions Rankings | ||
| Webometrics Ranking Web of Universities |

Omsk State Agrarian University n.a. P. A. Stolypin
- University rankings - Omsk State Agrarian University n.a. P. A. Stolypin
| RUR Academic Rankings | |||
| RUR Reputation Ranking | |||
| RUR World University Rankings | |||
| Show 2 more rankings of Omsk State Agrarian University n.a. P. A. Stolypin | |||

in Business, Computer Science, Medicine, Law, Education, Health... Study at your own pace , conveniently from home .

Omsk State Pedagogical University
- University rankings - Omsk State Pedagogical University
| RUR World University Rankings | |||
| RUR Reputation Ranking | |||
| RUR Academic Rankings | |||
| Show 1 more rankings of Omsk State Pedagogical University | |||

Omsk State Transport University
- University rankings - Omsk State Transport University
| Webometrics Ranking Web of Universities |
Omsk State University
- University rankings - Omsk State University
| Scimago Institutions Rankings | |||
| QS University Rankings: EECA Emerging Europe & Central Asia | |||
| QS University Rankings BRICS | |||
| Show 5 more rankings of Omsk State University | |||

Siberian State University of Physical Culture and Sports
Highest subject rankings of universities in Omsk
Omsk key facts for international students.
6 out of 7 Universities in Omsk Ranked in at least one ranking
10 Different Rankings list Universities in Omsk (9 institution and 1 subject rankings)
Population: 1173000
Time: GMT +6
District/province: Omsk,
* 100 = prices in London
- Living costs without accommodation 47* (53% cheaper than London)
- All costs including accommodation 65* (35% cheaper than London)
- Meals (grocery & lower cost restaurants) 34* (66% cheaper than London)
- Average Big Mac price 173.35 RUB
- Residential voltage: 230 V
- Frequency: 50 Hz
Map with location of universities in Omsk

Useful related pages

What is the best ranked university in Omsk?
What university in omsk is listed in most university rankings, what university in omsk is best ranked for studying engineering, ranking publishers, british quacquarelli symonds, uk, qs university rankings: eeca emerging europe & central asia (published: 15 december, 2021).
Academic reputation 30% Employer reputation 20% Faculty/student ratio 10% Papers per faculty 10% International research network 10%
view methodology
QS University Rankings BRICS (Published: 06 May, 2019)
Academic reputation 30% Employer reputation 20% Faculty/student ratio 20% Staff with a PhD 10% Papers per faculty 10%
RUR Ranking Agency (Moscow, Russia)
Rur world university rankings (published: 25 may, 2023).
Teaching: 40%
- Ratio Faculty/Student: 8%
- Ratio Faculty/Bachelor Degrees Awarded: 8%
- Ratio Faculty/Doctoral Degrees Awarded: 8%
- Ratio Doctoral Degrees Awarded/Bachelor Degrees Awarded: 8%
- World Teaching Reputation: 8%
Research: 40%
- Citations per Academic/Research Staff: 8%
- Doctoral Degrees per Accepted PhD: 8%
- Normalized Citation Impact: 8%
- Papers per Academic/Research Staff: 8%
- World Research Reputation: 8%
International Diversity: 10%
- International Faculty: 2%
- International Students: 2%
- International Co-Authored Papers: 2%
- Reputation Outside Geographical Region: 2%
- International Level: 2%
Financial Sustainability: 10%
- Institutional Income per Faculty: 2%
- Institutional Income per Student: 2%
- Papers per Research Income: 2%
- Research Income per Academic/Research Staff: 2%
- Research Income per Institutional Income: 2%
RUR Academic Rankings (Published: 25 May, 2023)
Normalized citation impact (Citations of research publications from all university authors compared with world averages) 20% Citation per papers 20% Papers per academic and research staff 20% International research reputation 20% Share of research publications written in international co-authorship 20%
RUR Reputation Ranking (Published: 25 May, 2023)
Teaching Reputation 50% Research Reputation 50%
Scimago Institutions
Scimago institutions rankings (published: 06 march, 2024).
Research 50% Innovation 30% Societal 20%
THE Times Higher Education, UK
The world university rankings (published: 27 september, 2023).
30% Teaching (the Learning Environment)
- Reputation survey: 15%
- Staff-to-student ratio: 4.5%
- Doctorate-to-bachelor’s ratio: 2.25%
- Doctorates-awarded-to-academic-staff ratio: 6%
- Institutional income: 2.25%
30% Research (Volume, Income and Reputation)
- Reputation survey: 18%
- Research income: 6%
- Research productivity: 6%
30% Citations (Research Influence)
7.5% International Outlook (Staff, Students and Research)
- Proportion of international students: 2.5%
- Proportion of international staff: 2.5%
- International collaboration: 2.5%
2.5% Industry Income (Knowledge Transfer)"
THE Emerging Economies University Ranking - Times Higher Education (Published: 19 October, 2021)
Teaching 30% Research (volume, income and reputation) 30% Citations 20% International outlook (staff, students, research) 10% Industry income (knowledge transfer) 10%
THE World University Impact Rankings - Overall (Published: 01 June, 2023)
Webometrics, webometrics ranking web of universities (published: 31 july, 2023).
Visibility 50% Excellence 35% Transparency 10% Presence 5%

- Biochemistry and Molecular Biology
- Biostatistics
- Environmental Health and Engineering
- Epidemiology
- Health Policy and Management
- Health, Behavior and Society
- International Health
- Mental Health
- Molecular Microbiology and Immunology
- Population, Family and Reproductive Health
- Program Finder
- Admissions Services
- Course Directory
- Academic Calendar
- Hybrid Campus
- Lecture Series
- Convocation
- Strategy and Development
- Implementation and Impact
- Integrity and Oversight
- In the School
- In the Field
- In Baltimore
- Resources for Practitioners
- Articles & News Releases
- In The News
- Statements & Announcements
- At a Glance
- Student Life
- Strategic Priorities
- Inclusion, Diversity, Anti-Racism, and Equity (IDARE)
- What is Public Health?
Concentration in Health Economics and Policy
Offered By: Department of Health Policy and Management
Onsite | Full-Time | 4 - 5 years
- MSPH Field Placements
- Master's Essays
- MAS Application Fee Waiver Requirements
- Master of Arts and Master of Science in Public Health (MA/MSPH)
- Master of Arts in Public Health Biology (MAPHB)
- Master of Bioethics (MBE)
- Mission, Vision, and Values
- Student Experience
- Program Outcomes
- For Hopkins Undergraduate Students
- Master of Health Science (MHS) - Department of Biochemistry and Molecular Biology
- Master of Health Science (MHS) - Department of Epidemiology
- Alumni Update
- MHS Combined with a Certificate Program
- Master of Health Science (MHS) - Department of Molecular Microbiology and Immunology
- Bachelor's/MHS in Health Economics and Outcomes Research
- MHS HEOR Careers
- Frequently Asked Questions
- Master of Health Science (MHS)
- Concurrent School-Wide Master of Health Science Program in Biostatistics
- Master of Health Science - Department of Population, Family and Reproductive Health
- Master of Health Science Online (MHS) - Department of Population, Family and Reproductive Health
- Careers in Health Economics
- Core Competencies
- Meet the Director
- What is Health Economics
- MPH Capstone Schedule
- Concentrations
- Online/Part-Time Format
- Requirements
Tuition and Funding
- Executive Board Faculty
- Master of Science (ScM) - Department of Biochemistry and Molecular Biology
- Master of Science (ScM) - Department of Biostatistics
- Master of Science (ScM) - Department of Epidemiology
- Master of Science (ScM) - Department of Molecular Microbiology and Immunology
- Bachelor's/MSPH in Health Policy
- FAQ for MSPH in Health Policy
- Field Placement Experience
- MSPH Capstone
- MSPH Practicum
- Required and Elective Courses
- Student Timeline
- Career Opportunities
- 38-Week Dietetics Practicum
- Completion Requirements
- MSPH/RD Program FAQ
- Program Goals
- Application Fee Waiver Requirements
- Doctor of Philosophy (PhD) - Department of Biostatistics
- Doctor of Philosophy (PhD) - Department of Epidemiology
- Program Goals and Expectations
- Doctor of Philosophy (PhD) - Department of Molecular Microbiology and Immunology
- Doctor of Philosophy (PhD) - Department of Population, Family and Reproductive Health
- Doctor of Philosophy (PhD) in Clinical Investigation
- Recent Graduates and Dissertation Titles
- PhD Funding
- PhD TA Requirement
- Recent Dissertation Titles
- JHU-Tsinghua Doctor of Public Health
- Prerequisites
- Concentration in Women’s and Reproductive Health
- Custom Track
- Concentration in Environmental Health
- Concentration in Global Health: Policy and Evaluation
- Concentration in Health Equity and Social Justice
- Concentration in Health Policy and Management
- Concentration in Implementation Science
- Combined Bachelor's / Master's Programs
- Concurrent MHS Option for BSPH Doctoral Students
- Concurrent MSPH Option for JHSPH Doctoral students
- Doctor of Medicine and Doctor of Philosophy (MD/PhD)
- Adolescent Health Certificate Program
- Bioethics Certificate Program
- Clinical Trials Certificate Program
- Community- Based Public Health Certificate Program
- Demographic Methods Certificate Program
- Epidemiology for Public Health Professionals Certificate Program
- Evaluation: International Health Programs Certificate Program
- Frequently Asked Questions for Certificate Programs
- Gender and Health Certificate Program
- Gerontology Certificate Program
- Global Digital Health Certificate Program
- Global Health Certificate Program
- Global Health Practice Certificate Program
- Health Communication Certificate Program
- Health Disparities and Health Inequality Certificate Program
- Health Education Certificate Program
- Health Finance and Management Certificate Program
- Health and Human Rights Certificate Program
- Healthcare Epidemiology and Infection Prevention and Control Certificate Program
- Humanitarian Health Certificate Program
- Implementation Science and Research Practice Certificate Program
- Injury and Violence Prevention Certificate Program
- International Healthcare Management and Leadership Certificate Program
- Leadership for Public Health and Healthcare Certificate Program
- Lesbian, Gay, Bisexual, Transgender, and Queer (LGBTQ) Public Health Certificate Program
- Maternal and Child Health Certificate Program
- Mental Health Policy, Economics and Services Certificate Program
- Non-Degree Students General Admissions Info
- Pharmacoepidemiology and Drug Safety Certificate Program
- Population Health Management Certificate Program
- Population and Health Certificate Program
- Public Health Advocacy Certificate Program
- Public Health Economics Certificate Program
- Public Health Informatics Certificate Program
- Public Health Practice Certificate Program
- Public Health Training Certificate for American Indian Health Professionals
- Public Mental Health Research Certificate Program
- Quality, Patient Safety and Outcomes Research Certificate Program
- Quantitative Methods in Public Health Certificate Program
- Requirements for Successful Completion of a Certificate Program
- Rigor, Reproducibility, and Responsibility in Scientific Practice Certificate Program
- Risk Sciences and Public Policy Certificate Program
- Spatial Analysis for Public Health Certificate Program
- Training Certificate in Public Health
- Tropical Medicine Certificate Program
- Tuition for Certificate Programs
- Vaccine Science and Policy Certificate Program
- Online Student Experience
- MAS and Affiliated Certificate Programs
- Barcelona Information
- Registration, Tuition, and Fees
- Agency Scholarship Application
- General Scholarship Application
- UPF Scholarship Application
- Course Evaluations
- Online Courses
- Registration
- General Institute Tuition Information
- International Students
- Directions to the Bloomberg School
- All Courses
- Important Guidance for ONSITE Students
- D.C. Courses
- Registration and Fees
- Cancellation and Closure Policies
- Application Procedures
- Career Search
- Current Activities
- Current Trainees
- Related Links
- Process for Appointing Postdoctoral Fellows
- Message from the Director
- Program Details
- Admissions FAQ
- Current Residents
- Elective Opportunities for Visiting Trainees
- What is Occupational and Environmental Medicine?
- Admissions Info
- Graduates by Year
- Compensation and Benefits
- How to Apply
- Academic Committee
- Course Details and Registration
- Tuition and Fees
- ONLINE SOCI PROGRAM
- Principal Faculty
- General Application
- JHHS Application
- Our Faculty
- Descripción los Cursos
- Programa en Epidemiología para Gestores de Salud, Basado en Internet
- Consultants
- Britt Dahlberg, PhD
- Joke Bradt, PhD, MT-BC
- Mark R. Luborsky, PhD
- Marsha Wittink, PhD
- Rebekka Lee, ScD
- Su Yeon Lee-Tauler, PhD
- Theresa Hoeft, PhD
- Vicki L. Plano Clark, PhD
- Program Retreat
- Mixed Methods Applications: Illustrations
- Announcements
- 2023 Call for Applications
- Jennifer I Manuel, PhD, MSW
- Joke Bradt, PhD
- Josiemer Mattei, PhD, MPH
- Justin Sanders, MD, MSc
- Linda Charmaran, PhD
- Nao Hagiwara, PhD
- Nynikka R. A. Palmer, DrPH, MPH
- Olayinka O. Shiyanbola, BPharm, PhD
- Sarah Ronis, MD, MPH
- Susan D. Brown, PhD
- Tara Lagu, MD, MPH
- Theresa Hoft, PhD
- Wynne E. Norton, PhD
- Yvonne Mensa-Wilmot, PhD, MPH
- A. Susana Ramírez, PhD, MPH
- Animesh Sabnis, MD, MSHS
- Autumn Kieber-Emmons, MD, MPH
- Benjamin Han, MD, MPH
- Brooke A. Levandowski, PhD, MPA
- Camille R. Quinn, PhD, AM, LCSW
- Justine Wu, MD, MPH
- Kelly Aschbrenner, PhD
- Kim N. Danforth, ScD, MPH
- Loreto Leiva, PhD
- Marie Brault, PhD
- Mary E. Cooley, PhD, RN, FAAN
- Meganne K. Masko, PhD, MT-BC/L
- PhuongThao D. Le, PhD, MPH
- Rebecca Lobb, ScD, MPH
- Allegra R. Gordon, ScD MPH
- Anita Misra-Hebert, MD MPH FACP
- Arden M. Morris, MD, MPH
- Caroline Silva, PhD
- Danielle Davidov, PhD
- Hans Oh, PhD
- J. Nicholas Dionne-Odom, PhD RN ACHPN
- Jacqueline Mogle, PhD
- Jammie Hopkins, DrPH, MS
- Joe Glass, PhD MSW
- Karen Whiteman, PhD MSW
- Katie Schultz, PhD MSW
- Rose Molina, MD
- Uriyoán Colón-Ramos, ScD MPA
- Andrew Riley, PhD
- Byron J. Powell, PhD, LCSW
- Carrie Nieman MD, MPH
- Charles R. Rogers, PhD, MPH, MS, CHES®
- Emily E. Haroz, PhD
- Jennifer Tsui, Ph.D., M.P.H.
- Jessica Magidson, PhD
- Katherine Sanchez, PhD, LCSW
- Kelly Doran, MD, MHS
- Kiara Alvarez, PhD
- LaPrincess C. Brewer, MD, MPH
- Melissa Radey, PhD, MA, MSSW
- Sophia L. Johnson, PharmD, MPH, PhD
- Supriya Gupta Mohile, MD, MS
- Virginia McKay, PhD
- Andrew Cohen, MD, PhD
- Angela Chen, PhD, PMHNP-BC, RN
- Christopher Salas-Wright, PhD, MSW
- Eliza Park MD, MS
- Jaime M. Hughes, PhD, MPH, MSW
- Johanne Eliacin, PhD, HSPP
- Lingrui Liu ScD MS
- Meaghan Kennedy, MD
- Nicole Stadnick, PhD, MPH
- Paula Aristizabal, MD
- Radhika Sundararajan, MD
- Sara Mamo, AuD, PhD
- Tullika Garg, MD MPH FACS
- Allison Magnuson, DO
- Ariel Williamson PhD, DBSM
- Benita Bamgbade, PharmD, PhD
- Christopher Woodrell MD
- Hung-Jui (Ray) Tan, MD, MSHPM
- Jasmine Abrams, PhD
- Jose Alejandro Rauh-Hain, MD
- Karen Flórez, DrPH, MPH
- Lavanya Vasudevan, PhD, MPH, CPH
- Maria Garcia, MD, MPH
- Robert Brady, PhD
- Saria Hassan, MD
- Scherezade Mama, DrPH
- Yuan Lu, ScD
- 2021 Scholars
- Sign Up for Our Email List
- Workforce Training
- Cells-to-Society Courses
- Course/Section Numbers Explained
- Pathway Program with Goucher College
- The George G. Graham Lecture
About the Concentration in Health Economics and Policy
The concentration in Health Economics and Policy prepares doctoral students to address the most pressing challenges in health and health care through innovative, rigorous and interdisciplinary research in the field of health economics. This program integrates traditional training in economics with practical training in health policy and health services research to train the next generation of health economists.
The curriculum offers a broad exposure to the health economics literature and public health disciplines, and stresses the policy implications of these fields of research. The curriculum stresses a foundation in applied modern microeconomic theory, economic evaluation, quantitative methods and econometrics, including PhD-level courses from the Department of Economics in the Krieger School of Arts and Sciences.
Doctoral students are paired with a faculty adviser from the Health Economics concentration with similar research interests. Faculty in the Health Economics concentration are working in a variety of research areas including understanding health insurance design, the economic implications of health and health care disparities, market forces and health care prices, pharmaceutical economics, and payment design and access. Doctoral students will also have the opportunity to work with other faculty within the Department, as well as faculty from other Departments including International Health, Population, Family, and Reproductive Health, Biostatistics, the School of Medicine, School of Nursing, the Carey Business School, and the Department of Economics. Students also often work with various centers and initiatives across the University, including the Hopkins Business of Health Initiative.
What Can You Do With a Graduate Degree In Health Economics And Policy?
The program prepares students for successful research careers as health economists. Former students have gone onto careers in academia, government, research-oriented non-profits, and the private sector. Visit the Graduate Employment Outcomes Dashboard to learn about Bloomberg School graduates' employment status, sector, and salaries.
View a list of selected recent graduates and dissertation titles for the PhD Concentration in Health Economics and Policy.
Curriculum for the Concentration in Health Economics and Policy
Browse an overview of the requirements for this PhD program in the JHU Academic Catalogue and explore all course offerings in the Bloomberg School Course Directory .
Admissions Requirements
For general admissions requirements, please visit the How to Apply page.
Standardized Test Scores
Standardized test scores are not required and not reviewed for this program. If you have taken a standardized test such as the GRE, GMAT, or MCAT and want to submit your scores, please note that they will not be used as a metric during the application review. Applications will be reviewed holistically based on all required application components.
Matthew Eisenberg, PhD, MPhil,
uses applied health economics methods to study how consumers make decisions about their healthcare.
Per the Collective Bargaining Agreement (CBA) with the JHU PhD Union, the minimum guaranteed 2025-2026 academic year stipend is $50,000 for all PhD students with a 4% increase the following year. Tuition, fees, and medical benefits are provided, including health insurance premiums for PhD student’s children and spouses of international students, depending on visa type. The minimum stipend and tuition coverage is guaranteed for at least the first four years of a BSPH PhD program; specific amounts and the number of years supported, as well as work expectations related to that stipend will vary across departments and funding source. Please refer to the CBA to review specific benefits, compensation, and other terms.
Need-Based Relocation Grants Students who are admitted to PhD programs at JHU starting in Fall 2023 or beyond can apply to receive a need-based grant to offset the costs of relocating to be able to attend JHU. These grants provide funding to a portion of incoming students who, without this money, may otherwise not be able to afford to relocate to JHU for their PhD program. This is not a merit-based grant. Applications will be evaluated solely based on financial need. View more information about the need-based relocation grants for PhD students .
Questions about the program? We're happy to help. [email protected]

IMAGES
COMMENTS
The PhD program in Public Health enhances commitment its PhD students who identify as underrepresented minority students, first-generation college graduates and students from economically disadvantaged backgrounds by offering research awards to the top candidates admitted to the program. Each year a minimum of two PhD admitted students will be ...
The PhD in Health Policy and Management is a full-time doctoral program that trains its students to conduct original investigator-initiated research through a combination of coursework and research mentoring. The curriculum includes core coursework that is common across the four concentrations and courses specific to each individual concentration.
The PhD program in epidemiology and clinical research will provide methodologic and interdisciplinary training that will equip students to carry out cutting-edge epidemiologic research. The program trains students in the tools of modern epidemiology, with heavy emphases on statistics, computer science, genetics, genomics, and bioinformatics.
The Doctor of Philosophy in Health Services and Policy Research (HSPR) program provides funding to all students admitted into the program for four years. These funding opportunities include obtaining research assistantship opportunities on various grant-funded projects with faculty in the Boston University School of Public Health, School of ...
Overview The PhD program in epidemiology and clinical research provides methodologic and interdisciplinary training to equip students to carry out cutting-edge epidemiologic research. The program trains students in the tools of modern epidemiology, with heavy emphasis on statistics, computer science, genetics, genomics, and bioinformatics.
Harvard PhD Program in Health Policy The Harvard PhD in Health Policy, awarded by the Harvard Kenneth C. Griffin Graduate School of Arts and Sciences, is a collaborative program among six Harvard University faculties: Faculty of Arts and Sciences, Harvard Business School, Harvard Kennedy School, Harvard Law School, Harvard Medical School, and Harvard T.H. Chan School of Public Health.
The Doctor of Philosophy in Health Services & Policy Research (PhD) degree program offered by the Department of Health Law, Policy & Management is designed to provide individuals with excellent research skills for use in academic, industry, or government settings. Students have the opportunity to collaborate with senior faculty in innovative research crucial to the improvement of healthcare ...
DescriptionThe PhD Program in Health Services trains health services researchers and health policy analysts for careers in academic institutions, health delivery systems, public health departments, government agencies, and the private sector. The program prepares students to conduct high-quality independent, collaborative research and policy ...
The PhD in Health Services Research and Health Policy at the Rollins School of Public Health at Emory University is a full-time program that trains researchers in the fields of health policy, health economics, health management, and health services research. Students take doctoral-level classes in the Department of Economics, the Department of ...
Since 1994, the PhD program in Health Services Research and Policy in the Department of Public Health Sciences has been highly successful in preparing students to become scholars for research, teaching, and public service careers in university, public policy, and governmental settings.
This Doctor of Philosophy (PhD) in Health Services Research and Policy program is designed to train doctoral students to pursue original quantitative, qualitative and/or mixed methods research in the broad areas of health policy and health services research in the United States and abroad, with an emphasis on research that can be translated ...
The PhD concentration in Health Services Research and Policy focuses on public health principles, research and evaluation methods, policy analysis, and areas related to health and health services delivery and population health.
The PhD in Health Services Research consists of a common core curriculum, concentration, and elective courses in either Health Systems and Policy or Knowledge Discovery and Health Informatics, and dissertation sequence courses.
The doctoral program in Epidemiology is anchored in public health and population research and analysis. Students approach research using epidemiologic methods to understand complex human health problems. The PhD requires two years of coursework followed by two (or more) years of research. Students are required to complete a teaching training ...
The Health Policy PhD program emphasizes the use of economics, health services and outcomes research to study today's challenging health policy issues.
The Health Systems PhD program equips students to become leaders in global health, conduct cutting-edge research, and design equitable health systems for underserved communities.
The PhD Program is unique among many health services and health policy oriented doctoral programs as it is designed with the inclusion of a Master of Science (MS) in Economics degree to be completed prior to entering the dissertation phase of the program.
The program trains researchers in the areas of health outcomes research, health services research and health data science to meet the changing needs of the health care system. This program is a dual-degree program that will grant the graduate with both an M.D. and Ph.D.
Health outcomes research is a rapidly expanding, interdisciplinary field that provides evidence and guidance for understanding the endpoints of treatments, interventions and health care practices, be they clinical, functional, quality-of-life or economic. Saint Louis University's Doctor of Philosophy (Ph.D.) in Health Outcomes Research is a program that trains researchers in the areas of ...
K. Davina Frick (she/her) is a professor who teaches economics for decision-making, business leadership and human values, frameworks for analyzing healthcare markets, and a course on how the U.S. health care system in the past, present, and future facilitates innovation.
Master's Degree or PhD in outcomes research, epidemiology, pharmacoepidemiology, public health, health services research or related quantitative or analytical field. Exposure to clinical, claims, pharmacy or other healthcare data. Additional Information. At Humana, we know your well-being is important to you, and it's important to us too.
In this Review, Pai and colleagues examine the global landscape of drug-resistant tuberculosis, exploring its epidemiology, causes, risk factors, stigma and associated mental health burden as well ...
Postdoctoral Research Associate in Medical Physics. ... Applicants will have a PhD or DSc degree in medical physics, biomedical engineering, radiological sciences, particle physics or other relevant engineering or physical sciences. ... diagnostic imaging, dosimetry, health physics, radiation safety, nuclear medicine, and imaging. Find a job ...
Marvin A Gonzalez Quiroz, MD, Ph.D., MSc, Assistant Professor in the Department of Environmental and Occupational Health in the UT School of Public Health San Antonio, has successfully secured a $30,000 grant from the Medical Research Council in the United Kingdom to support the 10th year of follow-up fieldwork for a community-based cohort study on chronic kid
Mycobacterium tuberculosis is an ancient bacterial pathogen that has acquired drug resistance to all drugs that have been used against it, despite lacking several key mechanisms available to other bacteria to facilitate rapid spread of resistance such as horizontal gene transfer and mobile resistance elements.
Innovative Projects, Research & Development. Youth Innovation Center. Scientific Society for Young Scientists and Students. OSMU KEY RESEARCH DIRECTIONS FOR 2022. 1. Quality management system of environment and population health: implementation at the regional level. 2. Patient-oriented technology implementation for treatment patients with ...
Universities in Omsk, Russia are listed in 10 rankings. All university rankings and student reviews in one place & explained. Student satisfaction, Academic reputation.
The concentration in Health Economics and Policy prepares doctoral students to address the most pressing challenges in health and health care through innovative, rigorous and interdisciplinary research in the field of health economics. This program integrates traditional training in economics with practical training in health policy and health ...