

Your Data Guide

How to Perform Hypothesis Testing Using Python

Step into the intriguing world of hypothesis testing, where your natural curiosity meets the power of data to reveal truths!
This article is your key to unlocking how those everyday hunches—like guessing a group’s average income or figuring out who owns their home—can be thoroughly checked and proven with data.
Thanks for reading Your Data Guide! Subscribe for free to receive new posts and support my work.
I am going to take you by the hand and show you, in simple steps, how to use Python to explore a hypothesis about the average yearly income.
By the time we’re done, you’ll not only get the hang of creating and testing hypotheses but also how to use statistical tests on actual data.
Perfect for up-and-coming data scientists, anyone with a knack for analysis, or just if you’re keen on data, get ready to gain the skills to make informed decisions and turn insights into real-world actions.
Join me as we dive deep into the data, one hypothesis at a time!
Before we get started, elevate your data skills with my expert eBooks—the culmination of my experiences and insights.
Support my work and enhance your journey. Check them out:

eBook 1: Personal INTERVIEW Ready “SQL” CheatSheet
eBook 2: Personal INTERVIEW Ready “Statistics” Cornell Notes
Best Selling eBook: Top 50+ ChatGPT Personas for Custom Instructions
Data Science Bundle ( Cheapest ): The Ultimate Data Science Bundle: Complete
ChatGPT Bundle ( Cheapest ): The Ultimate ChatGPT Bundle: Complete
💡 Checkout for more such resources: https://codewarepam.gumroad.com/
What is a hypothesis, and how do you test it?
A hypothesis is like a guess or prediction about something specific, such as the average income or the percentage of homeowners in a group of people.
It’s based on theories, past observations, or questions that spark our curiosity.
For instance, you might predict that the average yearly income of potential customers is over $50,000 or that 60% of them own their homes.
To see if your guess is right, you gather data from a smaller group within the larger population and check if the numbers ( like the average income, percentage of homeowners, etc. ) from this smaller group match your initial prediction.
You also set a rule for how sure you need to be to trust your findings, often using a 5% chance of error as a standard measure . This means you’re 95% confident in your results. — Level of Significance (0.05)
There are two main types of hypotheses : the null hypothesi s, which is your baseline saying there’s no change or difference, and the alternative hypothesis , which suggests there is a change or difference.
For example,
If you start with the idea that the average yearly income of potential customers is $50,000,
The alternative could be that it’s not $50,000—it could be less or more, depending on what you’re trying to find out.
To test your hypothesis, you calculate a test statistic —a number that shows how much your sample data deviates from what you predicted.
How you calculate this depends on what you’re studying and the kind of data you have. For example, to check an average, you might use a formula that considers your sample’s average, the predicted average, the variation in your sample data, and how big your sample is.
This test statistic follows a known distribution ( like the t-distribution or z-distribution ), which helps you figure out the p-value.
The p-value tells you the odds of seeing a test statistic as extreme as yours if your initial guess was correct.
A small p-value means your data strongly disagrees with your initial guess.
Finally, you decide on your hypothesis by comparing the p-value to your error threshold.
If the p-value is smaller or equal, you reject the null hypothesis, meaning your data shows a significant difference that’s unlikely due to chance.
If the p-value is larger, you stick with the null hypothesis , suggesting your data doesn’t show a meaningful difference and any change might just be by chance.
We’ll go through an example that tests if the average annual income of prospective customers exceeds $50,000.
This process involves stating hypotheses , specifying a significance level , collecting and analyzing data , and drawing conclusions based on statistical tests.
Example: Testing a Hypothesis About Average Annual Income
Step 1: state the hypotheses.
Null Hypothesis (H0): The average annual income of prospective customers is $50,000.
Alternative Hypothesis (H1): The average annual income of prospective customers is more than $50,000.
Step 2: Specify the Significance Level
Significance Level: 0.05, meaning we’re 95% confident in our findings and allow a 5% chance of error.
Step 3: Collect Sample Data
We’ll use the ProspectiveBuyer table, assuming it's a random sample from the population.
This table has 2,059 entries, representing prospective customers' annual incomes.
Step 4: Calculate the Sample Statistic
In Python, we can use libraries like Pandas and Numpy to calculate the sample mean and standard deviation.
SampleMean: 56,992.43
SampleSD: 32,079.16
SampleSize: 2,059
Step 5: Calculate the Test Statistic
We use the t-test formula to calculate how significantly our sample mean deviates from the hypothesized mean.
Python’s Scipy library can handle this calculation:
T-Statistic: 4.62
Step 6: Calculate the P-Value
The p-value is already calculated in the previous step using Scipy's ttest_1samp function, which returns both the test statistic and the p-value.
P-Value = 0.0000021
Step 7: State the Statistical Conclusion
We compare the p-value with our significance level to decide on our hypothesis:
Since the p-value is less than 0.05, we reject the null hypothesis in favor of the alternative.
Conclusion:
There’s strong evidence to suggest that the average annual income of prospective customers is indeed more than $50,000.
This example illustrates how Python can be a powerful tool for hypothesis testing, enabling us to derive insights from data through statistical analysis.
How to Choose the Right Test Statistics
Choosing the right test statistic is crucial and depends on what you’re trying to find out, the kind of data you have, and how that data is spread out.
Here are some common types of test statistics and when to use them:
T-test statistic:
This one’s great for checking out the average of a group when your data follows a normal distribution or when you’re comparing the averages of two such groups.
The t-test follows a special curve called the t-distribution . This curve looks a lot like the normal bell curve but with thicker ends, which means more chances for extreme values.
The t-distribution’s shape changes based on something called degrees of freedom , which is a fancy way of talking about your sample size and how many groups you’re comparing.
Z-test statistic:
Use this when you’re looking at the average of a normally distributed group or the difference between two group averages, and you already know the standard deviation for all in the population.
The z-test follows the standard normal distribution , which is your classic bell curve centered at zero and spreading out evenly on both sides.
Chi-square test statistic:
This is your go-to for checking if there’s a difference in variability within a normally distributed group or if two categories are related.
The chi-square statistic follows its own distribution, which leans to the right and gets its shape from the degrees of freedom —basically, how many categories or groups you’re comparing.
F-test statistic:
This one helps you compare the variability between two groups or see if the averages of more than two groups are all the same, assuming all groups are normally distributed.
The F-test follows the F-distribution , which is also right-skewed and has two types of degrees of freedom that depend on how many groups you have and the size of each group.
In simple terms, the test you pick hinges on what you’re curious about, whether your data fits the normal curve, and if you know certain specifics, like the population’s standard deviation.
Each test has its own special curve and rules based on your sample’s details and what you’re comparing.
Join my community of learners! Subscribe to my newsletter for more tips, tricks, and exclusive content on mastering Data Science & AI. — Your Data Guide Join my community of learners! Subscribe to my newsletter for more tips, tricks, and exclusive content on mastering data science and AI. By Richard Warepam ⭐️ Visit My Gumroad Shop: https://codewarepam.gumroad.com/
Ready for more?
Hypothesis Testing with Python
Learn how to plan, implement, and interpret different kinds of hypothesis tests in Python.
- AI assistance for guided coding help
- Projects to apply new skills
- Quizzes to test your knowledge
- A certificate of completion
Skill level
Time to complete
Prerequisites
About this course
In this course, you’ll learn to plan, implement, and interpret a hypothesis test in Python. Hypothesis testing is used to address questions about a population based on a subset from that population. For example, A/B testing is a framework for learning about consumer behavior based on a small sample of consumers.
This course assumes some preexisting knowledge of Python, including the NumPy and pandas libraries.
Introduction to Hypothesis Testing
Find out what you’ll learn in this course and why it’s important.
Hypothesis testing: Testing a Sample Statistic
Learn about hypothesis testing and implement binomial and one-sample t-tests in Python.
Hypothesis Testing: Testing an Association
Learn about hypothesis tests that can be used to evaluate whether there is an association between two variables.
Experimental Design
Learn to design an experiment to make a decision using a hypothesis test.
Hypothesis Testing Projects
Practice your hypothesis testing skills with some additional projects!
Certificate of completion available with Plus or Pro
The platform
Hands-on learning

Projects in this course
Heart disease research part i, heart disease research part ii, a/b testing at nosh mish mosh, earn a certificate of completion.
- Show proof Receive a certificate that demonstrates you've completed a course or path.
- Build a collection The more courses and paths you complete, the more certificates you collect.
- Share with your network Easily add certificates of completion to your LinkedIn profile to share your accomplishments.

Reviews from learners
Our learners work at.
- Google Logo
- Amazon Logo
- Microsoft Logo
- Reddit Logo
- Spotify Logo
- YouTube Logo
- Instagram Logo
Frequently asked questions about Hypothesis Testing with Python
What is hypothesis testing.
After drawing conclusions from data, you have to make sure it’s correct, and hypothesis testing involves using statistical methods to validate our results.
Why is hypothesis testing important?
What kind of jobs perform hypothesis testing, what else should i study if i am learning about hypothesis testing, join over 50 million learners and start hypothesis testing with python today, looking for something else, related resources, software testing methodologies, introduction to testing with mocha and chai, testing types, related courses and paths, hypothesis testing: associations, hypothesis testing: experimental design, browse more topics.
- Math 85,399 learners enrolled
- Python 3,443,842 learners enrolled
- Data Science 4,260,983 learners enrolled
- Code Foundations 7,095,668 learners enrolled
- Computer Science 5,544,604 learners enrolled
- Web Development 4,741,585 learners enrolled
- For Business 3,126,645 learners enrolled
- JavaScript 2,764,873 learners enrolled
- Data Analytics 2,261,059 learners enrolled

Unlock additional features with a paid plan
Practice projects, assessments, certificate of completion.
Statistical Hypothesis Testing: A Comprehensive Guide

We’ve all heard it – “ go to college to get a good job .” The assumption is that higher education leads straight to higher incomes. Elite Indian institutes like the IITs and IIMs are even judged based on the average starting salaries of their graduates. But is this direct connection between schooling and income actually true?
Intuitively, it seems believable. But how can we really prove this assumption that more school = more money? Is there hard statistical evidence either way? Turns out, there are methods to scientifically test widespread beliefs like this – what statisticians call hypothesis testing.
In this article, we’ll dig into the concept of hypothesis testing and the tools to rigorously question conventional wisdom: null and alternate hypotheses, one and two-tailed tests, paired sample tests, and more.
Statistical hypothesis testing allows researchers to make inferences about populations based on sample data. It involves setting up a null hypothesis, choosing a confidence level, calculating a p-value, and conducting tests such as two-tailed, one-tailed, or paired sample tests to draw conclusions.
What is Hypothesis Testing?
Statistical Hypothesis Testing is a method used to make inferences about a population based on sample data. Before we move ahead and understand what Hypothesis Testing is, we need to understand some basic terms.
Null Hypothesis
The Null Hypothesis is generally where we start our journey. Null Hypotheses are statements that are generally accepted or statements that you want to challenge. Since it is generally accepted that income level is positively correlated with quality of education, this will be our Null Hypothesis. It is denoted by H 0 .
H 0 : Income levels are positively correlated with quality of education.
Alternate Hypothesis
The Alternate Hypothesis is the opposite of the Null hypothesis. An alternate Hypothesis is what we want to prove as a researcher and is not generally accepted by society. An alternate hypothesis is denoted H a . The alternate hypothesis of the above is given below.
H a : Income levels are negatively correlated with the quality of education.
Confidence Level (1- α )
Confidence Levels represent the probability that the range of values contains the true parameter value. The most common confidence levels are 95% and 99%. It can be interpreted that our test is 95% accurate if our confidence level is 95%. It is denoted by 1-α.
p-value ( p )
The p-value represents the probability of obtaining test results at least as extreme as the results actually observed, under the assumption that the null hypothesis is correct. A lower p-value means fewer chances for our observed result to happen. If our p-value is less than α , our null hypothesis is rejected, otherwise null hypothesis is accepted.
Types of Hypothesis Tests
Since we are equipped with the basic terms, let’s go ahead and conduct some hypothesis tests.
Conducting a Two-Tailed Hypothesis Test
In a two-tailed hypothesis test, our analysis can go in either direction i.e. either more than or less than our observed value. For example, a medical researcher testing out the effects of a placebo wants to know whether it increases or decreases blood pressure. Let’s look at its Python implementation.
In the above code, we want to know if the group study method is an effective way to study or not. Therefore our null and alternate hypotheses are as follows.
- H 0 : The Group study method is not an effective way to study .
- H a : The group study method is an effective way to study .

Since the p-value is greater than α , we fail to reject the null hypothesis. Therefore the group study method is not an effective way to study.
Recommended: Hypothesis Testing in Python: Finding the critical value of T
In a one-tailed hypothesis test, we have certain expectations in which way our observed value will move i.e. higher or lower. For example, our researchers want to know if a particular medicine lowers our cholesterol level. Let’s look at its Python code.
Here our null and alternate hypothesis tests are given below.
- H 0 : The Group study method does not increase our marks.
- H a : The group study method increases our marks.

Since the p-value is greater than α , we fail to reject the null hypothesis. Therefore the group study method does not increase our marks.
A paired sample test compares two sets of observations and then provides us with a conclusion. For example, we need to know whether the reaction time of our participants increases after consuming caffeine. Let’s look at another example with a Python code as well.
Similar to the above hypothesis tests, we consider the group study method here as well. Our null and alternate hypotheses are as follows.
- H 0 : The group study method does not provide us with significant differences in our scores.
- H a : The group study method gives us significant differences in our scores.

Since the p-value is greater than α , we fail to reject the null hypothesis.
Here you go! Now you are equipped to perform statistical hypothesis testing on different samples and draw out different conclusions. You need to collect data and decide on null and alternate hypotheses. Furthermore, based on the predetermined hypothesis, you need to decide on which type of test to perform. Statistical hypothesis testing is one of the most powerful tools in the world of research.
Now that you have a grasp on statistical hypothesis testing, how will you apply these concepts to your own research or data analysis projects? What hypotheses are you eager to test?
Do check out: How to find critical value in Python
What Is Hypothesis Testing? Types and Python Code Example

Curiosity has always been a part of human nature. Since the beginning of time, this has been one of the most important tools for birthing civilizations. Still, our curiosity grows — it tests and expands our limits. Humanity has explored the plains of land, water, and air. We've built underwater habitats where we could live for weeks. Our civilization has explored various planets. We've explored land to an unlimited degree.
These things were possible because humans asked questions and searched until they found answers. However, for us to get these answers, a proven method must be used and followed through to validate our results. Historically, philosophers assumed the earth was flat and you would fall off when you reached the edge. While philosophers like Aristotle argued that the earth was spherical based on the formation of the stars, they could not prove it at the time.
This is because they didn't have adequate resources to explore space or mathematically prove Earth's shape. It was a Greek mathematician named Eratosthenes who calculated the earth's circumference with incredible precision. He used scientific methods to show that the Earth was not flat. Since then, other methods have been used to prove the Earth's spherical shape.
When there are questions or statements that are yet to be tested and confirmed based on some scientific method, they are called hypotheses. Basically, we have two types of hypotheses: null and alternate.
A null hypothesis is one's default belief or argument about a subject matter. In the case of the earth's shape, the null hypothesis was that the earth was flat.
An alternate hypothesis is a belief or argument a person might try to establish. Aristotle and Eratosthenes argued that the earth was spherical.
Other examples of a random alternate hypothesis include:
- The weather may have an impact on a person's mood.
- More people wear suits on Mondays compared to other days of the week.
- Children are more likely to be brilliant if both parents are in academia, and so on.
What is Hypothesis Testing?
Hypothesis testing is the act of testing whether a hypothesis or inference is true. When an alternate hypothesis is introduced, we test it against the null hypothesis to know which is correct. Let's use a plant experiment by a 12-year-old student to see how this works.
The hypothesis is that a plant will grow taller when given a certain type of fertilizer. The student takes two samples of the same plant, fertilizes one, and leaves the other unfertilized. He measures the plants' height every few days and records the results in a table.
After a week or two, he compares the final height of both plants to see which grew taller. If the plant given fertilizer grew taller, the hypothesis is established as fact. If not, the hypothesis is not supported. This simple experiment shows how to form a hypothesis, test it experimentally, and analyze the results.
In hypothesis testing, there are two types of error: Type I and Type II.
When we reject the null hypothesis in a case where it is correct, we've committed a Type I error. Type II errors occur when we fail to reject the null hypothesis when it is incorrect.
In our plant experiment above, if the student finds out that both plants' heights are the same at the end of the test period yet opines that fertilizer helps with plant growth, he has committed a Type I error.
However, if the fertilized plant comes out taller and the student records that both plants are the same or that the one without fertilizer grew taller, he has committed a Type II error because he has failed to reject the null hypothesis.
What are the Steps in Hypothesis Testing?
The following steps explain how we can test a hypothesis:
Step #1 - Define the Null and Alternative Hypotheses
Before making any test, we must first define what we are testing and what the default assumption is about the subject. In this article, we'll be testing if the average weight of 10-year-old children is more than 32kg.
Our null hypothesis is that 10 year old children weigh 32 kg on average. Our alternate hypothesis is that the average weight is more than 32kg. Ho denotes a null hypothesis, while H1 denotes an alternate hypothesis.
Step #2 - Choose a Significance Level
The significance level is a threshold for determining if the test is valid. It gives credibility to our hypothesis test to ensure we are not just luck-dependent but have enough evidence to support our claims. We usually set our significance level before conducting our tests. The criterion for determining our significance value is known as p-value.
A lower p-value means that there is stronger evidence against the null hypothesis, and therefore, a greater degree of significance. A p-value of 0.05 is widely accepted to be significant in most fields of science. P-values do not denote the probability of the outcome of the result, they just serve as a benchmark for determining whether our test result is due to chance. For our test, our p-value will be 0.05.
Step #3 - Collect Data and Calculate a Test Statistic
You can obtain your data from online data stores or conduct your research directly. Data can be scraped or researched online. The methodology might depend on the research you are trying to conduct.
We can calculate our test using any of the appropriate hypothesis tests. This can be a T-test, Z-test, Chi-squared, and so on. There are several hypothesis tests, each suiting different purposes and research questions. In this article, we'll use the T-test to run our hypothesis, but I'll explain the Z-test, and chi-squared too.
T-test is used for comparison of two sets of data when we don't know the population standard deviation. It's a parametric test, meaning it makes assumptions about the distribution of the data. These assumptions include that the data is normally distributed and that the variances of the two groups are equal. In a more simple and practical sense, imagine that we have test scores in a class for males and females, but we don't know how different or similar these scores are. We can use a t-test to see if there's a real difference.
The Z-test is used for comparison between two sets of data when the population standard deviation is known. It is also a parametric test, but it makes fewer assumptions about the distribution of data. The z-test assumes that the data is normally distributed, but it does not assume that the variances of the two groups are equal. In our class test example, with the t-test, we can say that if we already know how spread out the scores are in both groups, we can now use the z-test to see if there's a difference in the average scores.
The Chi-squared test is used to compare two or more categorical variables. The chi-squared test is a non-parametric test, meaning it does not make any assumptions about the distribution of data. It can be used to test a variety of hypotheses, including whether two or more groups have equal proportions.
Step #4 - Decide on the Null Hypothesis Based on the Test Statistic and Significance Level
After conducting our test and calculating the test statistic, we can compare its value to the predetermined significance level. If the test statistic falls beyond the significance level, we can decide to reject the null hypothesis, indicating that there is sufficient evidence to support our alternative hypothesis.
On the other contrary, if the test statistic does not exceed the significance level, we fail to reject the null hypothesis, signifying that we do not have enough statistical evidence to conclude in favor of the alternative hypothesis.
Step #5 - Interpret the Results
Depending on the decision made in the previous step, we can interpret the result in the context of our study and the practical implications. For our case study, we can interpret whether we have significant evidence to support our claim that the average weight of 10 year old children is more than 32kg or not.
For our test, we are generating random dummy data for the weight of the children. We'll use a t-test to evaluate whether our hypothesis is correct or not.
For a better understanding, let's look at what each block of code does.
The first block is the import statement, where we import numpy and scipy.stats . Numpy is a Python library used for scientific computing. It has a large library of functions for working with arrays. Scipy is a library for mathematical functions. It has a stat module for performing statistical functions, and that's what we'll be using for our t-test.
The weights of the children were generated at random since we aren't working with an actual dataset. The random module within the Numpy library provides a function for generating random numbers, which is randint .
The randint function takes three arguments. The first (20) is the lower bound of the random numbers to be generated. The second (40) is the upper bound, and the third (100) specifies the number of random integers to generate. That is, we are generating random weight values for 100 children. In real circumstances, these weight samples would have been obtained by taking the weight of the required number of children needed for the test.
Using the code above, we declared our null and alternate hypotheses stating the average weight of a 10-year-old in both cases.
t_stat and p_value are the variables in which we'll store the results of our functions. stats.ttest_1samp is the function that calculates our test. It takes in two variables, the first is the data variable that stores the array of weights for children, and the second (32) is the value against which we'll test the mean of our array of weights or dataset in cases where we are using a real-world dataset.
The code above prints both values for t_stats and p_value .
Lastly, we evaluated our p_value against our significance value, which is 0.05. If our p_value is less than 0.05, we reject the null hypothesis. Otherwise, we fail to reject the null hypothesis. Below is the output of this program. Our null hypothesis was rejected.
In this article, we discussed the importance of hypothesis testing. We highlighted how science has advanced human knowledge and civilization through formulating and testing hypotheses.
We discussed Type I and Type II errors in hypothesis testing and how they underscore the importance of careful consideration and analysis in scientific inquiry. It reinforces the idea that conclusions should be drawn based on thorough statistical analysis rather than assumptions or biases.
We also generated a sample dataset using the relevant Python libraries and used the needed functions to calculate and test our alternate hypothesis.
Thank you for reading! Please follow me on LinkedIn where I also post more data related content.
Read more posts .
If this article was helpful, share it .
Learn to code for free. freeCodeCamp's open source curriculum has helped more than 40,000 people get jobs as developers. Get started
Next-Gen App & Browser Testing Cloud
Trusted by 2 Mn+ QAs & Devs to accelerate their release cycles
Automation Playwright Testing Selenium Python Tutorial
- What Is Hypothesis Testing in Python: A Hands-On Tutorial
Jaydeep Karale
Posted On: June 5, 2024
In software testing, there is an approach known as property-based testing that leverages the concept of formal specification of code behavior and focuses on asserting properties that hold true for a wide range of inputs rather than individual test cases.
Python is an open-source programming language that provides a Hypothesis library for property-based testing. Hypothesis testing in Python provides a framework for generating diverse and random test data, allowing development and testing teams to thoroughly test their code against a broad spectrum of inputs.
In this blog, we will explore the fundamentals of Hypothesis testing in Python using Selenium and Playwright. We’ll learn various aspects of Hypothesis testing, from basic usage to advanced strategies, and demonstrate how it can improve the robustness and reliability of the codebase.
TABLE OF CONTENTS
What Is a Hypothesis Library?
Decorators in hypothesis, strategies in hypothesis, setting up python environment for hypothesis testing, how to perform hypothesis testing in python, hypothesis testing in python with selenium and playwright.
- How to Run Hypothesis Testing in Python With Date Strategy?
- How to Write Composite Strategies in Hypothesis Testing in Python?
Frequently Asked Questions (FAQs)
Hypothesis is a property-based testing library that automates test data generation based on properties or invariants defined by the developers and testers.
In property-based testing, instead of specifying individual test cases, developers define general properties that the code should satisfy. Hypothesis then generates a wide range of input data to test these properties automatically.
Property-based testing using Hypothesis allows developers and testers to focus on defining the behavior of their code rather than writing specific test cases, resulting in more comprehensive testing coverage and the discovery of edge cases and unexpected behavior.
Writing property-based tests usually consists of deciding on guarantees our code should make – properties that should always hold, regardless of what the world throws at the code.
Examples of such guarantees can be:
- Your code shouldn’t throw an exception or should only throw a particular type of exception (this works particularly well if you have a lot of internal assertions).
- If you delete an object, it is no longer visible.
- If you serialize and then deserialize a value, you get the same value back.
Before we proceed further, it’s worthwhile to understand decorators in Python a bit since the Hypothesis library exposes decorators that we need to use to write tests.
In Python, decorators are a powerful feature that allows you to modify or extend the behavior of functions or classes without changing their source code. Decorators are essentially functions themselves, which take another function (or class) as input and return a new function (or class) with added functionality.
Decorators are denoted by the @ symbol followed by the name of the decorator function placed directly before the definition of the function or class to be modified.
Let us understand this with the help of an example:

In the example above, only authenticated users are allowed to create_post() . The logic to check authentication is wrapped in its own function, authenticate() .
This function can now be called using @authenticate before beginning a function where it’s needed & Python would automatically know that it needs to execute the code of authenticate() before calling the function.
If we no longer need the authentication logic in the future, we can simply remove the @authenticate line without disturbing the core logic. Thus, decorators are a powerful construct in Python that allows plug-n-play of repetitive logic into any function/method.
Now that we know the concept of Python decorators, let us understand the given decorators that which Hypothesis provides.
Hypothesis @given Decorator
This decorator turns a test function that accepts arguments into a randomized test. It serves as the main entry point to the Hypothesis.
The @given decorator can be used to specify which arguments of a function should be parameterized over. We can use either positional or keyword arguments, but not a mixture of both.
| .given(*_given_arguments, **_given_kwargs) |
Some valid declarations of the @given decorator are:
| given(integers(), integers()) a(x, y): pass given(integers()) b(x, y): pass given(y=integers()) c(x, y): pass given(x=integers()) d(x, y): pass given(x=integers(), y=integers()) e(x, **kwargs): pass given(x=integers(), y=integers()) f(x, *args, **kwargs): pass SomeTest(TestCase): @given(integers()) def test_a_thing(self, x): pass |
Some invalid declarations of @given are:
| given(integers(), integers(), integers()) g(x, y): pass given(integers()) h(x, *args): pass given(integers(), x=integers()) i(x, y): pass given() j(x, y): pass |
Hypothesis @example Decorator
When writing production-grade applications, the ability of a Hypothesis to generate a wide range of input test data plays a crucial role in ensuring robustness.
However, there are certain inputs/scenarios the testing team might deem mandatory to be tested as part of every test run. Hypothesis has the @example decorator in such cases where we can specify values we always want to be tested. The @example decorator works for all strategies.
Let’s understand by tweaking the factorial test example.

The above test will always run for the input value 41 along with other custom-generated test data by the Hypothesis st.integers() function.
By now, we understand that the crux of the Hypothesis is to test a function for a wide range of inputs. These inputs are generated automatically, and the Hypothesis lets us configure the range of inputs. Under the hood, the strategy method takes care of the process of generating this test data of the correct data type.
Hypothesis offers a wide range of strategies such as integers, text, boolean, datetime, etc. For more complex scenarios, which we will see a bit later in this blog, the hypothesis also lets us set up composite strategies.
While not exhaustive, here is a tabular summary of strategies available as part of the Hypothesis library.
| Strategy | Description |
|---|---|
| Generates none values. | |
| Generates boolean values (True or False). | |
| Generates integer values. | |
| Generates floating-point values. | |
| Generates unicode text strings. | |
| Generates single unicode characters. | |
| Generates lists of elements. | |
| Generates tuples of elements. | |
| Generates dictionaries with specified keys and values. | |
| Generates sets of elements. | |
| Generates binary data. | |
| Generates datetime objects. | |
| Generates timedelta objects. | |
| Choose one of the given strategies with equal probability. | |
| Chooses values from a given sequence with equal probability. | |
| Generates lists of elements. | |
| Generates date objects. | |
| Generates datetime objects. | |
| Generates a single value. | |
| Generates strings that match a given regular expression. | |
| Generates UUID objects. | |
| Generates complex numbers. | |
| Generates fraction objects. | |
| Builds objects using a provided constructor and strategy for each argument. | |
| Generates single unicode characters. | |
| Generates unicode text strings. | |
| Chooses values from a given sequence with equal probability. | |
| Generates arbitrary data values. | |
| Generates values that are shared between different parts of a test. | |
| Generates recursively structured data. | |
| Generates data based on the outcome of other strategies. |
Let’s see the steps to how to set up a test environment to perform Hypothesis testing in Python.
- Create a separate virtual environment for this project using the built-in venv module of Python using the command.

- Activate the newly created virtual environment using the activate script present within the environment.

- Install the Hypothesis library necessary for property-based testing using the pip install hypothesis command. The installed package can be viewed using the command pip list. When writing this blog, the latest version of Hypothesis is 6.102.4. For this article, we have used the Hypothesis version 6.99.6.

- Install python-dotenv , pytest, Playwright, and Selenium packages which we will need to run the tests on the cloud. We will talk about this in more detail later in the blog.
Our final project structure setup looks like below:
With the setup done, let us now understand Hypothesis testing in Python with various examples, starting with the introductory one and then working toward more complex ones.
Subscribe to the LambdaTest YouTube Channel for quick updates on the tutorials around Selenium Python and more.
Let’s now start writing tests to understand how we can leverage the Hypothesis library to perform Python automation .
For this, let’s look at one test scenario to understand Hypothesis testing in Python.
Test Scenario:
Implementation:
This is what the initial implementation of the function looks like:
| factorial(num: int) -> int: if num < 0: raise ValueError("Input must be > 0") fact = 1 for _ in range(1, num + 1): fact *= _ return fact |
It takes in an integer as an input. If the input is 0, it raises an error; if not, it uses the range() function to generate a list of numbers within, iterate over it, calculate the factorial, and return it.
Let’s now write a test using the Hypothesis library to test the above function:
| hypothesis import given, strategies as st given(st.integers(min_value=1, max_value=30)) test_factorial(num: int): fact_num_result = factorial(num) fact_num_minus_one_result = factorial(num-1) result = fact_num_result / fact_num_minus_one_result assert num == result |
Code Walkthrough:
Let’s now understand the step-by-step code walkthrough for Hypothesis testing in Python.
Step 1: From the Hypothesis library, we import the given decorator and strategies method.

Step 2: Using the imported given and strategies, we set our test strategy of passing integer inputs within the range of 1 to 30 to the function under test using the min_value and max_value arguments.

Step 3: We write the actual test_factorial where the integer generated by our strategy is passed automatically by Hypothesis into the value num.
Using this value we call the factorial function once for value num and num – 1.
Next, we divide the factorial of num by the factorial of num -1 and assert if the result of the operation is equal to the original num.

Test Execution:
Let’s now execute our hypothesis test using the pytest -v -k “test_factorial” command.

And Hypothesis confirms that our function works perfectly for the given set of inputs, i.e., for integers from 1 to 30.
We can also view detailed statistics of the Hypothesis run by passing the argument –hypothesis-show-statistics to pytest command as:
| -v --hypothesis-show-statistics -k "test_factorial" |

The difference between the reuse and generate phase in the output above is explained below:
- Reuse Phase: During the reuse phase, the Hypothesis attempts to reuse previously generated test data. If a test case fails or raises an exception, the Hypothesis will try to shrink the failing example to find a minimal failing case.
This phase typically has a very short runtime, as it involves reusing existing test data or shrinking failing examples. The output provides statistics about the typical runtimes and the number of passing, failing, and invalid examples encountered during this phase.
- Generate Phase: During the generate phase, the Hypothesis generates new test data based on the defined strategies. This phase involves generating a wide range of inputs to test the properties defined by the developer.
The output provides statistics about the typical runtimes and the number of passing, failing, and invalid examples generated during this phase. While this helped us understand what passing tests look like with a Hypothesis, it’s also worthwhile to understand how a Hypothesis can catch bugs in the code.
Let’s rewrite the factorial() function with an obvious bug, i.e., remove the check for when the input value is 0.
| factorial(num: int) -> int: # if num < 0: # raise ValueError("Number must be >= 0") fact = 1 for _ in range(1, num + 1): fact *= _ return fact |
We also tweak the test to remove the min_value and max_value arguments.
| given(st.integers()) test_factorial(num: int): fact_num_result = factorial(num) fact_num_minus_one_result = factorial(num-1) result = int(fact_num_result / fact_num_minus_one_result) assert num == result |
Let us now rerun the test with the same command:
| -v --hypothesis-show-statistics -k test_factorial |
| pytest -v --hypothesis-show-statistics -k test_factorial |
We can clearly see how Hypothesis has caught the bug immediately, which is shown in the above output. Hypothesis presents the input that resulted in the failing test under the Falsifying example section of the output.

So far, we’ve performed Hypothesis testing locally. This works nicely for unit tests , but when setting up automation for building more robust and resilient test suites, we can leverage a cloud grid like LambdaTest that supports automation testing tools like Selenium and Playwright.
LambdaTest is an AI-powered test orchestration and execution platform that enables developers and testers to perform automation testing with Selenium and Playwright at scale. It provides a remote test lab of 3000+ real environments.
How to Perform Hypothesis Testing in Python Using Cloud Selenium Grid?
Selenium is an open-source suite of tools and libraries for web automation . When combined with a cloud grid, it can help you perform Hypothesis testing in Python with Selenium at scale.
Let’s look at one test scenario to understand Hypothesis testing in Python with Selenium.
The code to set up a connection to LambdaTest Selenium Grid is stored in a crossbrowser_selenium.py file.
| selenium import webdriver selenium.webdriver.chrome.options import Options selenium.webdriver.common.keys import Keys time import sleep urllib3 warnings os selenium.webdriver import ChromeOptions selenium.webdriver import FirefoxOptions selenium.webdriver.remote.remote_connection import RemoteConnection hypothesis.strategies import integers dotenv import load_dotenv () = os.getenv('LT_USERNAME', None) = os.getenv('LT_ACCESS_KEY', None) CrossBrowserSetup: global web_driver def __init__(self): global remote_url urllib3.disable_warnings(urllib3.exceptions.InsecureRequestWarning) remote_url = "https://" + str(username) + ":" + str(access_key) + "@hub.lambdatest.com/wd/hub" def add(self, browsertype): if (browsertype == "Firefox"): ff_options = webdriver.FirefoxOptions() ff_options.browser_version = "latest" ff_options.platform_name = "Windows 11" lt_options = {} lt_options["build"] = "Build: FF: Hypothesis Testing with Selenium & Pytest" lt_options["project"] = "Project: FF: Hypothesis Testing withSelenium & Pytest" lt_options["name"] = "Test: FF: Hypothesis Testing with Selenium & Pytest" lt_options["browserName"] = "Firefox" lt_options["browserVersion"] = "latest" lt_options["platformName"] = "Windows 11" lt_options["console"] = "error" lt_options["w3c"] = True lt_options["headless"] = False ff_options.set_capability('LT:Options', lt_options) web_driver = webdriver.Remote( command_executor = remote_url, options = ff_options ) self.driver = web_driver self.driver.get("https://www.lambdatest.com") sleep(1) if web_driver is not None: web_driver.execute_script("lambda-status=passed") web_driver.quit() return True else: return False |
The test_selenium.py contains code to test the Hypothesis that tests will only run on the Firefox browser.
| hypothesis import given, settings hypothesis import given, example hypothesis.strategies as strategy src.crossbrowser_selenium import CrossBrowserSetup settings(deadline=None) given(strategy.just("Firefox")) test_add(browsertype_1): cbt = CrossBrowserSetup() assert True == cbt.add(browsertype_1) |
Let’s now understand the step-by-step code walkthrough for Hypothesis testing in Python using Selenium Grid.
Step 1: We import the necessary Selenium methods to initiate a connection to LambdaTest Selenium Grid.
The FirefoxOptions() method is used to configure the setup when connecting to LambdaTest Selenium Grid using Firefox.

Step 2: We use the load_dotenv package to access the LT_ACCESS_KEY required to access the LambdaTest Selenium Grid, which is stored in the form of environment variables.

The LT_ACCESS_KEY can be obtained from your LambdaTest Profile > Account Settings > Password & Security .

Step 3: We initialize the CrossBrowserSetup class, which prepares the remote connection URL using the username and access_key.

Step 4: The add() method is responsible for checking the browsertype and then setting the capabilities of the LambdaTest Selenium Grid.

LambdaTest offers a variety of capabilities, such as cross browser testing , which means we can test on various operating systems such as Windows, Linux, and macOS and multiple browsers such as Chrome, Firefox, Edge, and Safari.
For the purpose of this blog, we will be testing that connection to the LambdaTest Selenium Grid should only happen if the browsertype is Firefox.
Step 5: If the connection to LambdaTest happens, the add() returns True ; else, it returns False .

Let’s now understand a step-by-step walkthrough of the test_selenium.py file.
Step 1: We set up the imports of the given decorator and the Hypothesis strategy. We also import the CrossBrowserSetup class.

Step 2: @setting(deadline=None) ensures the test doesn’t timeout if the connection to the LambdaTest Grid takes more time.
We use the @given decorator to set the strategy to just use Firefox as an input to the test_add() argument broswertype_1. We then initialize an instance of the CrossBrowserSetup class & call the add() method using the broswertype_1 & assert if it returns True .
The commented strategy @given(strategy.just(‘Chrome’)) is to demonstrate that the add() method, when called with Chrome, returns False .

Let’s now run the test using pytest -k “test_hypothesis_selenium.py”.

We can see that the test has passed, and the Web Automation Dashboard reflects that the connection to the Selenium Grid has been successful.

On opening one of the execution runs, we can see a detailed step-by-step test execution.

How to Perform Hypothesis Testing in Python Using Cloud Playwright Grid?
Playwright is a popular open-source tool for end-to-end testing developed by Microsoft. When combined with a cloud grid, it can help you perform Hypothesis testing in Python at scale.
Let’s look at one test scenario to understand Hypothesis testing in Python with Playwright.
| website. |
| os dotenv import load_dotenv playwright.sync_api import expect, sync_playwright hypothesis import given, strategies as st subprocess urllib json () = { 'browserName': 'Chrome', # Browsers allowed: `Chrome`, `MicrosoftEdge`, `pw-chromium`, `pw-firefox` and `pw-webkit` 'browserVersion': 'latest', 'LT:Options': { 'platform': 'Windows 11', 'build': 'Playwright Hypothesis Demo Build', 'name': 'Playwright Locators Test For Windows 11 & Chrome', 'user': os.getenv('LT_USERNAME'), 'accessKey': os.getenv('LT_ACCESS_KEY'), 'network': True, 'video': True, 'visual': True, 'console': True, 'tunnel': False, # Add tunnel configuration if testing locally hosted webpage 'tunnelName': '', # Optional 'geoLocation': '', # country code can be fetched from https://www.lambdatest.com/capabilities-generator/ } interact_with_lambdatest(quantity): with sync_playwright() as playwright: playwrightVersion = str(subprocess.getoutput('playwright --version')).strip().split(" ")[1] capabilities['LT:Options']['playwrightClientVersion'] = playwrightVersion lt_cdp_url = 'wss://cdp.lambdatest.com/playwright?capabilities=' + urllib.parse.quote(json.dumps(capabilities)) browser = playwright.chromium.connect(lt_cdp_url) page = browser.new_page() page.goto("https://ecommerce-playground.lambdatest.io/") page.get_by_role("button", name="Shop by Category").click() page.get_by_role("link", name="MP3 Players").click() page.get_by_role("link", name="HTC Touch HD HTC Touch HD HTC Touch HD HTC Touch HD").click() page.get_by_role("button", name="Add to Cart").click(click_count=quantity) page.get_by_role("link", name="Checkout ").first.click() unit_price = float(page.get_by_role("cell", name="$146.00").first.inner_text().replace("$","")) page.evaluate("_ => {}", "lambdatest_action: {\"action\": \"setTestStatus\", \"arguments\": {\"status\":\"" + "Passed" + "\", \"remark\": \"" + "pass" + "\"}}" ) page.close() total_price = quantity * unit_price return total_price = st.integers(min_value=1, max_value=10) given(quantity=quantity_strategy) test_website_interaction(quantity): assert interact_with_lambdatest(quantity) == quantity * 146.00 |
Let’s now understand the step-by-step code walkthrough for Hypothesis testing in Python using Playwright Grid.
Step 1: To connect to the LambdaTest Playwright Grid, we need a Username and Access Key, which can be obtained from the Profile page > Account Settings > Password & Security.
We use the python-dotenv module to load the Username and Access Key, which are stored as environment variables.
The capabilities dictionary is used to set up the Playwright Grid on LambdaTest.
We configure the Grid to use Windows 11 and the latest version of Chrome.

Step 3: The function interact_with_lambdatest interacts with the LambdaTest eCommerce Playground website to simulate adding a product to the cart and proceeding to checkout.
It starts a Playwright session and retrieves the version of the Playwright being used. The LambdaTest CDP URL is created with the appropriate capabilities. It connects to the Chromium browser instance on LambdaTest.
A new page instance is created, and the LambdaTest eCommerce Playground website is navigated. The specified product is added to the cart by clicking through the required buttons and links. The unit price of the product is extracted from the web page.
The browser page is then closed.

Step 4: We define a Hypothesis strategy quantity_strategy using st.integers to generate random integers representing product quantities. The generated integers range from 1 to 10
Using the @given decorator from the Hypothesis library, we define a property-based test function test_website_interaction that takes a quantity parameter generated by the quantity_strategy .
Inside the test function, we use the interact_with_lambdatest function to simulate interacting with the website and calculate the total price based on the generated quantity.
We assert that the total_price returned by interact_with_lambdatest matches the expected value calculated as quantity * 146.00.

Let’s now run the test on the Playwright Cloud Grid using pytest -v -k “test_hypothesis_playwright.py ”

The LambdaTest Web Automation Dashboard shows successfully passed tests.

Run Your Hypothesis Tests With Selenium & Playwright on Cloud. Try LambdaTest Today!
How to Perform Hypothesis Testing in Python With Date Strategy?
In the previous test scenario, we saw a simple example where we used the integer() strategy available as part of the Hypothesis. Let’s now understand another strategy, the date() strategy, which can be effectively used to test date-based functions.
Also, the output of the Hypothesis run can be customized to produce detailed results. Often, we may wish to see an even more verbose output when executing a Hypothesis test.
To do so, we have two options: either use the @settings decorator or use the –hypothesis-verbosity=<verbosity_level> when performing pytest testing .
| hypothesis import Verbosity,settings, given, strategies as st datetime import datetime, timedelta generate_expiry_alert(expiry_date): current_date = datetime.now().date() days_until_expiry = (expiry_date - current_date).days return days_until_expiry <= 45 given(expiry_date=st.dates()) settings(verbosity=Verbosity.verbose, max_examples=1000) test_expiry_alert_generation(expiry_date): alert_generated = generate_expiry_alert(expiry_date) # Check if the alert is generated correctly based on the expiry date days_until_expiry = (expiry_date - datetime.now().date()).days expected_alert = days_until_expiry <= 45 assert alert_generated == expected_alert |
Let’s now understand the code step-by-step.
Step 1: The function generate_expiry_alert() , which takes in an expiry_date as input and returns a boolean depending on whether the difference between the current date and expiry_date is less than or equal to 45 days.

Step 2: To ensure we test the generate_expiry_alert() for a wide range of date inputs, we use the date() strategy.
We also enable verbose logging and set the max_examples=1000 , which requests Hypothesis to generate 1000 date inputs at the max.

Step 3: On the inputs generated by Hypothesis in Step 3, we call the generate_expiry_alert() function and store the returned boolean in alert_generated.
We then compare the value returned by the function generate_expiry_alert() with a locally calculated copy and assert if the match.

We execute the test using the below command in the verbose mode, which allows us to see the test input dates generated by the Hypothesis.
| -s --hypothesis-show-statistics --hypothesis-verbosity=debug -k "test_expiry_alert_generation" |

As we can see, Hypothesis ran 1000 tests, 2 with reused data and 998 with unique newly generated data, and found no issues with the code.
Now, imagine the trouble we would have had to take to write 1000 tests manually using traditional example-based testing.
How to Perform Hypothesis Testing in Python With Composite Strategies?
So far, we’ve been using simple standalone examples to demo the power of Hypothesis. Let’s now move on to more complicated scenarios.
| website offers customer rewards points. A class tracks the customer reward points and their spending. class. |
The implementation of the UserRewards class is stored in a user_rewards.py file for better readability.
| UserRewards: def __init__(self, initial_points): self.reward_points = initial_points def get_reward_points(self): return self.reward_points def spend_reward_points(self, spent_points): if spent_points<= self.reward_points: self.reward_points -= spent_points return True else: return False |
The tests for the UserRewards class are stored in test_user_rewards.py .
| hypothesis import given, strategies as st src.user_rewards import UserRewards = st.integers(min_value=0, max_value=1000) given(initial_points=reward_points_strategy) test_get_reward_points(initial_points): user_rewards = UserRewards(initial_points) assert user_rewards.get_reward_points() == initial_points given(initial_points=reward_points_strategy, spend_amount=st.integers(min_value=0, max_value=1000)) test_spend_reward_points(initial_points, spend_amount): user_rewards = UserRewards(initial_points) remaining_points = user_rewards.get_reward_points() if spend_amount <= initial_points: assert user_rewards.spend_reward_points(spend_amount) remaining_points -= spend_amount else: assert not user_rewards.spend_reward_points(spend_amount) assert user_rewards.get_reward_points() == remaining_points |
Let’s now understand what is happening with both the class file and the test file step-by-step, starting first with the UserReward class.
Step 1: The class takes in a single argument initial_points to initialize the object.

Step 2: The get_reward_points() function returns the customers current reward points.

Step 3: The spend_reward_points() takes in the spent_points as input and returns True if spent_points are less than or equal to the customer current point balance and updates the customer reward_points by subtracting the spent_points , else it returns False .

That is it for our simple UserRewards class. Next, we understand what’s happening in the test_user_rewards.py step-by-step.
Step 1: We import the @given decorator and strategies from Hypothesis and the UserRewards class.

Step 2: Since reward points will always be integers, we use the integer() Hypothesis strategy to generate 1000 sample inputs starting with 0 and store them in a reward_points_strategy variable.

Step 3: Use the rewards_point_strategy as an input we run the test_get_reward_points() for 1000 samples starting with value 0.
For each input, we initialize the UserRewards class and assert that the method get_reward_points() returns the same value as the initial_points .
Step 4: To test the spend_reward_points() function, we generate two sets of sample inputs first, an initial reward_points using the reward_points_strategy we defined in Step 2 and a spend_amount which simulates spending of points.

Step 5: Write the test_spend_reward_points , which takes in the initial_points and spend_amount as arguments and initializes the UserRewards class with initial_point .
We also initialize a remaining_points variable to track the points remaining after the spend.

Step 6: If the spend_amount is less than the initial_points allocated to the customer, we assert if spend_reward_points returns True and update the remaining_points else, we assert spend_reward_points returns False .

Step 7: Lastly, we assert if the final remaining_points are correctly returned by get_rewards_points , which should be updated after spending the reward points.

Let’s now run the test and see if Hypothesis is able to find any bugs in the code.
| -s --hypothesis-show-statistics --hypothesis-verbosity=debug -k "test_user_rewards" |

To test if the Hypothesis indeed works, let’s make a small change to the UserRewards by commenting on the logic to deduct the spent_points in the spend_reward_points() function.

We run the test suite again using the command pytest -s –hypothesis-show-statistics -k “test_user_rewards “.

This time, the Hypothesis highlights the failures correctly.
Thus, we can catch any bugs and potential side effects of code changes early, making it perfect for unit testing and regression testing .
To understand composite strategies a bit more, let’s now test the shopping cart functionality and see how composite strategy can help write robust tests for even the most complicated of real-world scenarios.
| and which handles the shopping cart feature of the website. |
Let’s view the implementation of the ShoppingCart class written in the shopping_cart.py file.
| random enum import Enum, auto Item(Enum): """Item type""" LUNIX_CAMERA = auto() IMAC = auto() HTC_TOUCH = auto() CANNON_EOS = auto() IPOD_TOUCH = auto() APPLE_VISION_PRO = auto() COFMACBOOKFEE = auto() GALAXY_S24 = auto() def __str__(self): return self.name.upper() ShoppingCart: def __init__(self): """ "" self.items = {} def add_item(self, item: Item, price: int | float, quantity: int = 1) -> None: """ "" if item.name in self.items: self.items[item.name]["quantity"] += quantity else: self.items[item.name] = {"price": price, "quantity": quantity} def remove_item(self, item: Item, quantity: int = 1) -> None: """ "" if item.name in self.items: if self.items[item.name]["quantity"] <= quantity: del self.items[item.name] else: self.items[item.name]["quantity"] -= quantity def get_total_price(self): total_price = 0 for item in self.items.values(): total_price += item["price"] * item["quantity"] return total_price |
Let’s now view the tests written to verify the correct behavior of all aspects of the ShoppingCart class stored in a separate test_shopping_cart.py file.
| typing import Callable hypothesis import given, strategies as st hypothesis.strategies import SearchStrategy src.shopping_cart import ShoppingCart, Item st.composite items_strategy(draw: Callable[[SearchStrategy[Item]], Item]): return draw(st.sampled_from(list(Item))) st.composite price_strategy(draw: Callable[[SearchStrategy[int]], int]): return draw(st.integers(min_value=1, max_value=100)) st.composite qty_strategy(draw: Callable[[SearchStrategy[int]], int]): return draw(st.integers(min_value=1, max_value=10)) given(items_strategy(), price_strategy(), qty_strategy()) test_add_item_hypothesis(item, price, quantity): cart = ShoppingCart() # Add items to cart cart.add_item(item=item, price=price, quantity=quantity) # Assert that the quantity of items in the cart is equal to the number of items added assert item.name in cart.items assert cart.items[item.name]["quantity"] == quantity given(items_strategy(), price_strategy(), qty_strategy()) test_remove_item_hypothesis(item, price, quantity): cart = ShoppingCart() print("Adding Items") # Add items to cart cart.add_item(item=item, price=price, quantity=quantity) cart.add_item(item=item, price=price, quantity=quantity) print(cart.items) # Remove item from cart print(f"Removing Item {item}") quantity_before = cart.items[item.name]["quantity"] cart.remove_item(item=item) quantity_after = cart.items[item.name]["quantity"] # Assert that if we remove an item, the quantity of items in the cart is equal to the number of items added - 1 assert quantity_before == quantity_after + 1 given(items_strategy(), price_strategy(), qty_strategy()) test_calculate_total_hypothesis(item, price, quantity): cart = ShoppingCart() # Add items to cart cart.add_item(item=item, price=price, quantity=quantity) cart.add_item(item=item, price=price, quantity=quantity) # Remove item from cart cart.remove_item(item=item) # Calculate total total = cart.get_total_price() assert total == cart.items[item.name]["price"] * cart.items[item.name]["quantity"] |
Code Walkthrough of ShoppingCart class:
Let’s now understand what is happening in the ShoppingCart class step-by-step.
Step 1: We import the Python built-in Enum class and the auto() method.
The auto function within the Enum module automatically assigns sequential integer values to enumeration members, simplifying the process of defining enumerations with incremental values.
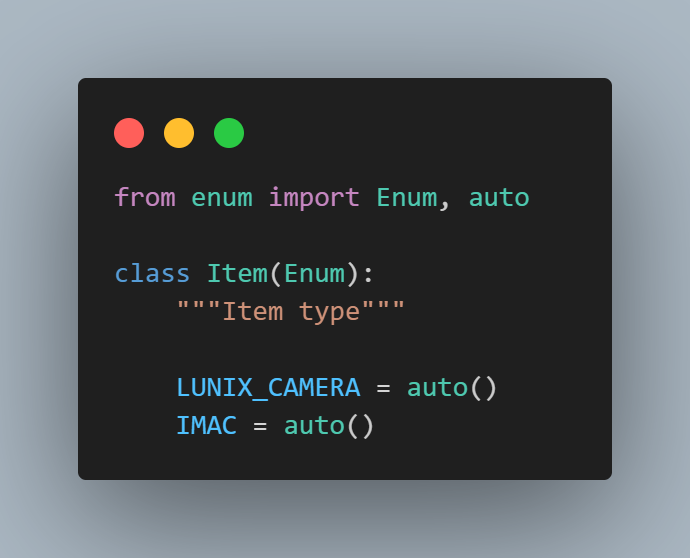
We define an Item enum corresponding to items available for sale on the LambdaTest eCommerce Playground website.
Step 2: We initialize the ShoppingCart class with an empty dictionary of items.

Step 3: The add_item() method takes in the item, price, and quantity as input and adds it to the shopping cart state held in the item dictionary.

Step 4: The remove_item() method takes in an item and quantity and removes it from the shopping cart state indicated by the item dictionary.

Step 5: The get_total_price() method iterates over the item dictionary, multiples the quantity by price, and returns the total_price of items in the cart.

Code Walkthrough of test_shopping_cart:
Let’s now understand step-by-step the tests written to ensure the correct working of the ShoppingCart class.
Step 1: First, we set up the imports, including the @given decorator, strategies, and the ShoppingCart class and Item enum.
The SearchStrategy is one of the various strategies on offer as part of the Hypothesis. It represents a set of rules for generating valid inputs to test a specific property or behavior of a function or program.

Step 2: We use the @st.composite decorator to define a custom Hypothesis strategy named items_strategy. This strategy takes a single argument, draw, which is a callable used to draw values from other strategies.
The st.sampled_from strategy randomly samples values from a given iterable. Within the strategy, we use draw(st.sampled_from(list(Item))) to draw a random Item instance from a list of all enum members.
Each time the items_strategy is used in a Hypothesis test, it will generate a random instance of the Item enum for testing purposes.

Step 3: The price_strategy runs on similar logic as the item_strategy but generates an integer value between 1 and 100.

Step 4: The qty_strategy runs on similar logic as the item_strategy but generates an integer value between 1 and 10.

Step 5: We use the @given decorator from the Hypothesis library to define a property-based test.
The items_strategy() , price_strategy() , and qty_strategy() functions are used to generate random values for the item, price, and quantity parameters, respectively.
Inside the test function, we create a new instance of a ShoppingCart .
We then add an item to the cart using the generated values for item, price, and quantity.
Finally, we assert that the item was successfully added to the cart and that the quantity matches the generated quantity.

Step 6: We use the @given decorator from the Hypothesis library to define a property-based test.
The items_strategy(), price_strategy() , and qty_strategy() functions are used to generate random values for the item, price, and quantity parameters, respectively.
Inside the test function, we create a new instance of a ShoppingCart . We then add the same item to the cart twice to simulate two quantity additions to the cart.
We remove one instance of the item from the cart. After that, we compare the item quantity before and after removal to ensure it decreases by 1.
The test verifies the behavior of the remove_item() method of the ShoppingCart class by testing it with randomly generated inputs for item, price , and quantity.

Step 7: We use the @given decorator from the Hypothesis library to define a property-based test.
The items_strategy(), price_strategy(), and qty_strategy() functions are used to generate random values for the item, price, and quantity parameters, respectively.
We add the same item to the cart twice to ensure it’s present, then remove one instance of the item from the cart. After that, we calculate the total price of items remaining in the cart.
Finally, we assert that the total price matches the price of one item times its remaining quantity.
The test verifies the correctness of the get_total_price() method of the ShoppingCart class by testing it with randomly generated inputs for item, price , and quantity .
Let’s now run the test using the command pytest –hypothesis-show-statistics -k “test_shopping_cart”.

We can verify that Hypothesis was able to find no issues with the ShoppingCart class.
Let’s now amend the price_strategy and qty_strategy to remove the min_value and max_value arguments.

And rerun the test pytest -k “test_shopping_cart” .

The tests run clearly reveal that we have bugs with respect to handling scenarios when quantity and price are passed as 0.
This also reveals the fact that setting the test inputs correctly to ensure we do comprehensive testing is key to writing robots and resilient tests.
Choosing min_val and max_val should only be done when we know beforehand the bounds of inputs the function under test will receive. If we are unsure what the inputs are, maybe it’s important to come up with the right strategies based on the behavior of the function under test.
In this blog we have seen in detail how Hypothesis testing in Python works using the popular Hypothesis library. Hypothesis testing falls under property-based testing and is much better than traditional testing in handling edge cases.
We also explored Hypothesis strategies and how we can use the @composite decorator to write custom strategies for testing complex functionalities.
We also saw how Hypothesis testing in Python can be performed with popular test automation frameworks like Selenium and Playwright. In addition, by performing Hypothesis testing in Python with LambdaTest on Cloud Grid, we can set up effective automation tests to enhance our confidence in the code we’ve written.
What are the three types of Hypothesis tests?
There are three main types of hypothesis tests based on the direction of the alternative hypothesis:
- Right-tailed test: This tests if a parameter is greater than a certain value.
- Left-tailed test: This tests if a parameter is less than a certain value.
- Two-tailed test: This tests for any non-directional difference, either greater or lesser than the hypothesized value.
What is Hypothesis testing in the ML model?
Hypothesis testing is a statistical approach used to evaluate the performance and validity of machine learning models. It helps us determine if a pattern observed in the training data likely holds true for unseen data (generalizability).
Jaydeep is a software engineer with 10 years of experience, most recently developing and supporting applications written in Python. He has extensive with shell scripting and is also an AI/ML enthusiast. He is also a tech educator, creating content on Twitter, YouTube, Instagram, and LinkedIn. Link to his YouTube channel- https://www.youtube.com/@jaydeepkarale
See author's profile
Author’s Profile
Got Questions? Drop them on LambdaTest Community. Visit now
Test Your Web Or Mobile Apps On 3000+ Browsers
Related Articles
How To Write Test Cases Effectively: Your Step-by-step Guide
September 5, 2024
Manual Testing | Automation |

How to Use Assert and Verify in Selenium
Faisal Khatri
September 2, 2024
Automation | Tutorial |

How To Download & Upload Files Using Selenium With Java
August 29, 2024
Automation | Selenium Tutorial | Tutorial |

Introducing KaneAI – World’s First End-to-End Testing Assistant
August 21, 2024
Automation | LambdaTest Updates |

ExpectedConditions In Selenium: Types And Examples
August 13, 2024

CSS Grid Best Practices: Guide With Examples
Tahera Alam
Web Development | LambdaTest Experiments | Tutorial |
Try LambdaTest Now !!
Get 100 minutes of automation test minutes FREE!!
Download Whitepaper
You'll get your download link by email.
Don't worry, we don't spam!
We use cookies to give you the best experience. Cookies help to provide a more personalized experience and relevant advertising for you, and web analytics for us. Learn More in our Cookies policy , Privacy & Terms of service .
Schedule Your Personal Demo ×
Pytest with Eric
How to Use Hypothesis and Pytest for Robust Property-Based Testing in Python
There will always be cases you didn’t consider, making this an ongoing maintenance job. Unit testing solves only some of these issues.
Example-Based Testing vs Property-Based Testing
Project set up, getting started, prerequisites.
| | |
Simple Example
Source code.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 | def find_largest_smallest_item(input_array: list) -> tuple: """ Function to find the largest and smallest items in an array :param input_array: Input array :return: Tuple of largest and smallest items """ if len(input_array) == 0: raise ValueError # Set the initial values of largest and smallest to the first item in the array largest = input_array[0] smallest = input_array[0] # Iterate through the array for i in range(1, len(input_array)): # If the current item is larger than the current value of largest, update largest if input_array[i] > largest: largest = input_array[i] # If the current item is smaller than the current value of smallest, update smallest if input_array[i] return largest, smallest def sort_array(input_array: list, sort_key: str) -> list: """ Function to sort an array :param sort_key: Sort key :param input_array: Input array :return: Sorted array """ if len(input_array) == 0: raise ValueError if sort_key not in input_array[0]: raise KeyError if not isinstance(input_array[0][sort_key], int): raise TypeError sorted_data = sorted(input_array, key=lambda x: x[sort_key], reverse=True) return sorted_data |
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 | def reverse_string(input_string) -> str: """ Function to reverse a string :param input_string: Input string :return: Reversed string """ return input_string[::-1] def complex_string_operation(input_string: str) -> str: """ Function to perform a complex string operation :param input_string: Input string :return: Transformed string """ # Remove Whitespace input_string = input_string.strip().replace(" ", "") # Convert to Upper Case input_string = input_string.upper() # Remove vowels vowels = ("A", "E", "I", "O", "U") for x in input_string.upper(): if x in vowels: input_string = input_string.replace(x, "") return input_string |
Simple Example — Unit Tests
Example-based testing.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 | pytest import logging from src.random_operations import ( reverse_string, find_largest_smallest_item, complex_string_operation, sort_array, ) logging.basicConfig(level=logging.INFO) logger = logging.getLogger(__name__) # Example Based Unit Testing def test_find_largest_smallest_item(): assert find_largest_smallest_item([1, 2, 3]) == (3, 1) def test_reverse_string(): assert reverse_string("hello") == "olleh" def test_sort_array(): data = [ {"name": "Alice", "age": 25}, {"name": "Bob", "age": 30}, {"name": "Charlie", "age": 20}, {"name": "David", "age": 35}, ] assert sort_array(data, "age") == [ {"name": "David", "age": 35}, {"name": "Bob", "age": 30}, {"name": "Alice", "age": 25}, {"name": "Charlie", "age": 20}, ] def test_complex_string_operation(): assert complex_string_operation(" Hello World ") == "HLLWRLD" |
Running The Unit Test
Property-based testing.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 | hypothesis import given, strategies as st from hypothesis import assume as hypothesis_assume # Property Based Unit Testing @given(st.lists(st.integers(), min_size=1, max_size=25)) def test_find_largest_smallest_item_hypothesis(input_list): assert find_largest_smallest_item(input_list) == (max(input_list), min(input_list)) @given( st.lists( st.fixed_dictionaries({"name": st.text(), "age": st.integers()}), )) def test_sort_array_hypothesis(input_list): if len(input_list) == 0: with pytest.raises(ValueError): sort_array(input_list, "age") hypothesis_assume(len(input_list) > 0) assert sort_array(input_list, "age") == sorted( input_list, key=lambda x: x["age"], reverse=True ) @given(st.text()) def test_reverse_string_hypothesis(input_string): assert reverse_string(input_string) == input_string[::-1] @given(st.text()) def test_complex_string_operation_hypothesis(input_string): assert complex_string_operation(input_string) == input_string.strip().replace( " ", "" ).upper().replace("A", "").replace("E", "").replace("I", "").replace( "O", "" ).replace( "U", "" ) |
Complex Example
Source code.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 | random from enum import Enum, auto class Item(Enum): """Item type""" APPLE = auto() ORANGE = auto() BANANA = auto() CHOCOLATE = auto() CANDY = auto() GUM = auto() COFFEE = auto() TEA = auto() SODA = auto() WATER = auto() def __str__(self): return self.name.upper() class ShoppingCart: def __init__(self): """ Creates a shopping cart object with an empty dictionary of items """ self.items = {} def add_item(self, item: Item, price: int | float, quantity: int = 1) -> None: """ Adds an item to the shopping cart :param quantity: Quantity of the item :param item: Item to add :param price: Price of the item :return: None """ if item.name in self.items: self.items[item.name]["quantity"] += quantity else: self.items[item.name] = {"price": price, "quantity": quantity} def remove_item(self, item: Item, quantity: int = 1) -> None: """ Removes an item from the shopping cart :param quantity: Quantity of the item :param item: Item to remove :return: None """ if item.name in self.items: if self.items[item.name]["quantity"] self.items[item.name] else: self.items[item.name]["quantity"] -= quantity def get_total_price(self): total = 0 for item in self.items.values(): total += item["price"] * item["quantity"] return total def view_cart(self) -> None: """ Prints the contents of the shopping cart :return: None """ print("Shopping Cart:") for item, price in self.items.items(): print("- {}: ${}".format(item, price)) def clear_cart(self) -> None: """ Clears the shopping cart :return: None """ self.items = {} |
Complex Example — Unit Tests
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 | pytest from src.shopping_cart import ShoppingCart, Item @pytest.fixture() def cart(): return ShoppingCart() # Define Items apple = Item.APPLE orange = Item.ORANGE gum = Item.GUM soda = Item.SODA water = Item.WATER coffee = Item.COFFEE tea = Item.TEA # Example Based Testing def test_add_item(cart): cart.add_item(apple, 1.00) cart.add_item(orange, 1.00) cart.add_item(gum, 2.00) cart.add_item(soda, 2.50) assert cart.items == { "APPLE": {"price": 1.0, "quantity": 1}, "ORANGE": {"price": 1.0, "quantity": 1}, "GUM": {"price": 2.0, "quantity": 1}, "SODA": {"price": 2.5, "quantity": 1}, } def test_remove_item(cart): cart.add_item(orange, 1.00) cart.add_item(tea, 3.00) cart.add_item(coffee, 3.00) cart.remove_item(orange) assert cart.items == { "TEA": {"price": 3.0, "quantity": 1}, "COFFEE": {"price": 3.0, "quantity": 1}, } def test_total(cart): cart.add_item(orange, 1.00) cart.add_item(apple, 2.00) cart.add_item(soda, 2.00) cart.add_item(soda, 2.00) cart.add_item(water, 1.00) cart.remove_item(apple) cart.add_item(gum, 2.50) assert cart.get_total_price() == 8.50 def test_clear_cart(cart): cart.add_item(apple, 1.00) cart.add_item(soda, 2.00) cart.add_item(water, 1.00) cart.clear_cart() assert cart.items == {} |
| 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 | typing import Callable from hypothesis import given, strategies as st from hypothesis.strategies import SearchStrategy from src.shopping_cart import ShoppingCart, Item # Create a strategy for items @st.composite def items_strategy(draw: Callable[[SearchStrategy[Item]], Item]): return draw(st.sampled_from(list(Item))) # Create a strategy for price @st.composite def price_strategy(draw: Callable[[SearchStrategy[float]], float]): return round(draw(st.floats(min_value=0.01, max_value=100, allow_nan=False)), 2) # Create a strategy for quantity @st.composite def qty_strategy(draw: Callable[[SearchStrategy[int]], int]): return draw(st.integers(min_value=1, max_value=10)) @given(items_strategy(), price_strategy(), qty_strategy()) def test_add_item_hypothesis(item, price, quantity): cart = ShoppingCart() # Add items to cart cart.add_item(item=item, price=price, quantity=quantity) # Assert that the quantity of items in the cart is equal to the number of items added assert item.name in cart.items assert cart.items[item.name]["quantity"] == quantity @given(items_strategy(), price_strategy(), qty_strategy()) def test_remove_item_hypothesis(item, price, quantity): cart = ShoppingCart() print("Adding Items") # Add items to cart cart.add_item(item=item, price=price, quantity=quantity) cart.add_item(item=item, price=price, quantity=quantity) print(cart.items) # Remove item from cart print(f"Removing Item {item}") quantity_before = cart.items[item.name]["quantity"] cart.remove_item(item=item) quantity_after = cart.items[item.name]["quantity"] # Assert that if we remove an item, the quantity of items in the cart is equal to the number of items added - 1 assert quantity_before == quantity_after + 1 @given(items_strategy(), price_strategy(), qty_strategy()) def test_calculate_total_hypothesis(item, price, quantity): cart = ShoppingCart() # Add items to cart cart.add_item(item=item, price=price, quantity=quantity) cart.add_item(item=item, price=price, quantity=quantity) # Remove item from cart cart.remove_item(item=item) # Calculate total total = cart.get_total_price() assert total == cart.items[item.name]["price"] * cart.items[item.name]["quantity"] |
Discover Bugs With Hypothesis
Define your own hypothesis strategies.
| 2 3 4 5 6 7 8 9 10 11 12 13 14 | @st.composite def items_strategy(draw: Callable[[SearchStrategy[Item]], Item]): return draw(st.sampled_from(list(Item))) # Create a strategy for price @st.composite def price_strategy(draw: Callable[[SearchStrategy[float]], float]): return round(draw(st.floats(min_value=0.01, max_value=100, allow_nan=False)), 2) # Create a strategy for quantity @st.composite def qty_strategy(draw: Callable[[SearchStrategy[int]], int]): return draw(st.integers(min_value=1, max_value=10)) |
Model-Based Testing in Hypothesis
Additional reading.
- Subscription
Hypothesis Testing in Python
In this course, you’ll learn advanced statistical concepts like significance testing and multi-category chi-square testing, which will help you perform more powerful and robust data analysis.
Part of the Data Analyst (Python) , and Data Scientist (Python) paths.
- Intermediate friendly

“Dataquest teaches you how you do data analysis in the real world. Dataquest is well-suited for folks with busier schedules. The lessons are broken into smaller chunks, so it’s really conducive to that. You only have an hour, okay, well, just do one or two lessons and that’s fine.”
Stacey Ustian
Course overview.
In this course, you’ll learn about single and multi-category chi-square tests, degrees of freedom, hypothesis testing, and different statistical distributions.
To learn about hypothesis testing and statistical significance, you’ll work hands-on with multiple datasets on weight loss data — are patients losing weight due to pure luck, or is it a diet pill? You’ll run the numbers and find out!
At the end of the course, you’ll complete a guided project in which you’ll work with data from the American TV show Jeopardy. You’ll analyze text and search for winning strategies. It’s a chance for you to combine the skills you learned in this course, and to showcase a fascinating project in your portfolio. Best of all, you’ll learn by doing — you’ll practice and get feedback directly in the browser.
- Defining regular and multi-category chi-squared tests
- Performing significance testing to understand an outcome's importance
Course outline
Hypothesis testing in python [4 lessons], significance testing 1h.
- Explain how hypothesis testing works
- Define the relation between statistical significance and hypothesis testing
Chi-Squared Tests 1h
- Determine the statistical significance of a set of categorical values
- Generate the chi-squared distribution
- Define degrees of freedom
Multi-Category Chi-Squared Tests 1h
- Extend chi-squared tests to multiple categories
- Calculate the statistical significance of multi-category chi-squared tests
Guided Project: Winning Jeopardy 1h
- Answer questions using text data
- Apply chi-squared tests to real problems
Projects in this course
Winning jeopardy.
For this project, you’ll take on the role of a Jeopardy contestant looking for any edge to win. You’ll work with a dataset of 20,000 Jeopardy questions using Python and pandas to analyze question and answer text and uncover helpful patterns.
The Dataquest guarantee
Dataquest has helped thousands of people start new careers in data. If you put in the work and follow our path, you’ll master data skills and grow your career.
We believe so strongly in our paths that we offer a full satisfaction guarantee. If you complete a career path on Dataquest and aren’t satisfied with your outcome, we’ll give you a refund.
Master skills faster with Dataquest
Go from zero to job-ready.
Learn exactly what you need to achieve your goal. Don’t waste time on unrelated lessons.

Build your project portfolio
Build confidence with our in-depth projects, and show off your data skills.
Challenge yourself with exercises
Work with real data from day one with interactive lessons and hands-on exercises.
Showcase your path certification
Share the evidence of your hard work with your network and potential employers.
Grow your career with Dataquest.

Aaron Melton
Business analyst at aditi consulting.
“Dataquest starts at the most basic level, so a beginner can understand the concepts. I tried learning to code before, using Codecademy and Coursera. I struggled because I had no background in coding, and I was spending a lot of time Googling. Dataquest helped me actually learn.”

Machine Learning Engineer at Twitter
“I liked the interactive environment on Dataquest. The material was clear and well organized. I spent more time practicing then watching videos and it made me want to keep learning.”

Victoria E. Guzik
Associate data scientist at callisto media.
“I really love learning on Dataquest. I looked into a couple of other options and I found that they were much too handhold-y and fill in the blank relative to Dataquest’s method. The projects on Dataquest were key to getting my job. I doubled my income!”
Join 1M+ data learners on Dataquest.
Create a free account, choose a learning path, complete exercises and projects, advance your career, start learning today.
- Quick start guide
- Edit on GitHub
Quick start guide ¶
This document should talk you through everything you need to get started with Hypothesis.
An example ¶
Suppose we’ve written a run length encoding system and we want to test it out.
We have the following code which I took straight from the Rosetta Code wiki (OK, I removed some commented out code and fixed the formatting, but there are no functional modifications):
We want to write a test for this that will check some invariant of these functions.
The invariant one tends to try when you’ve got this sort of encoding / decoding is that if you encode something and then decode it then you get the same value back.
Let’s see how you’d do that with Hypothesis:
(For this example we’ll just let pytest discover and run the test. We’ll cover other ways you could have run it later).
The text function returns what Hypothesis calls a search strategy. An object with methods that describe how to generate and simplify certain kinds of values. The @given decorator then takes our test function and turns it into a parametrized one which, when called, will run the test function over a wide range of matching data from that strategy.
Anyway, this test immediately finds a bug in the code:
Hypothesis correctly points out that this code is simply wrong if called on an empty string.
If we fix that by just adding the following code to the beginning of our encode function then Hypothesis tells us the code is correct (by doing nothing as you’d expect a passing test to).
If we wanted to make sure this example was always checked we could add it in explicitly by using the @example decorator:
This can be useful to show other developers (or your future self) what kinds of data are valid inputs, or to ensure that particular edge cases such as "" are tested every time. It’s also great for regression tests because although Hypothesis will remember failing examples , we don’t recommend distributing that database.
It’s also worth noting that both @example and @given support keyword arguments as well as positional. The following would have worked just as well:
Suppose we had a more interesting bug and forgot to reset the count each time. Say we missed a line in our encode method:
Hypothesis quickly informs us of the following example:
Note that the example provided is really quite simple. Hypothesis doesn’t just find any counter-example to your tests, it knows how to simplify the examples it finds to produce small easy to understand ones. In this case, two identical values are enough to set the count to a number different from one, followed by another distinct value which should have reset the count but in this case didn’t.
Installing ¶
Hypothesis is available on PyPI as “hypothesis” . You can install it with:
You can install the dependencies for optional extensions with e.g. pip install hypothesis[pandas,django] .
If you want to install directly from the source code (e.g. because you want to make changes and install the changed version), check out the instructions in CONTRIBUTING.rst .
Running tests ¶
In our example above we just let pytest discover and run our tests, but we could also have run it explicitly ourselves:
We could also have done this as a unittest.TestCase :
A detail: This works because Hypothesis ignores any arguments it hasn’t been told to provide (positional arguments start from the right), so the self argument to the test is simply ignored and works as normal. This also means that Hypothesis will play nicely with other ways of parameterizing tests. e.g it works fine if you use pytest fixtures for some arguments and Hypothesis for others.
Writing tests ¶
A test in Hypothesis consists of two parts: A function that looks like a normal test in your test framework of choice but with some additional arguments, and a @given decorator that specifies how to provide those arguments.
Here are some other examples of how you could use that:
Note that as we saw in the above example you can pass arguments to @given either as positional or as keywords.
Where to start ¶
You should now know enough of the basics to write some tests for your code using Hypothesis. The best way to learn is by doing, so go have a try.
If you’re stuck for ideas for how to use this sort of test for your code, here are some good starting points:
Try just calling functions with appropriate arbitrary data and see if they crash. You may be surprised how often this works. e.g. note that the first bug we found in the encoding example didn’t even get as far as our assertion: It crashed because it couldn’t handle the data we gave it, not because it did the wrong thing.
Look for duplication in your tests. Are there any cases where you’re testing the same thing with multiple different examples? Can you generalise that to a single test using Hypothesis?
This piece is designed for an F# implementation , but is still very good advice which you may find helps give you good ideas for using Hypothesis.
If you have any trouble getting started, don’t feel shy about asking for help .
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu
- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
A comparative study of sentiment classification models for greek reviews.

1. Introduction
2. theoretical background and review, 2.1. text representation, 2.2. computational methods for sentiment classification, 2.3. related research for greek sentiment classification, 3. methodology, 3.1. dataset selection, 3.2. text preprocessing, 3.3. modeling experiments, 3.3.1. machine learning approaches, 3.3.2. artficial neural network models, 3.3.3. transfer learning models, 3.3.4. large language models, 3.4. model evaluation, 4.1. machine learning, 4.2. artificial neural networks, 4.3. transfer learning model, 4.4. large language models, 5. discussion, 6. conclusions, data availability statement, acknowledgments, conflicts of interest.
- Nandwani, P.; Verma, R. A Review on Sentiment Analysis and Emotion Detection from Text. Soc. Netw. Anal. Min. 2021 , 11 , 81. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Al Maruf, A.; Khanam, F.; Haque, M.M.; Jiyad, Z.M.; Mridha, M.F.; Aung, Z. Challenges and Opportunities of Text-Based Emotion Detection: A Survey. IEEE Access 2024 , 12 , 18416–18450. [ Google Scholar ] [ CrossRef ]
- Zhang, W.; Li, X.; Deng, Y.; Bing, L.; Lam, W. A Survey on Aspect-Based Sentiment Analysis: Tasks, Methods, and Challenges. IEEE Trans. Knowl. Data Eng. 2023 , 35 , 11019–11038. [ Google Scholar ] [ CrossRef ]
- Li, R.; Chen, H.; Feng, F.; Ma, Z.; Wang, X.; Hovy, E. DualGCN: Exploring Syntactic and Semantic Information for Aspect-Based Sentiment Analysis. IEEE Trans. Neural Netw. Learn. Syst. 2024 , 35 , 7642–7656. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Alaei, A.R.; Becken, S.; Stantic, B. Sentiment Analysis in Tourism: Capitalizing on Big Data. J. Travel. Res. 2019 , 58 , 175–191. [ Google Scholar ] [ CrossRef ]
- Alamoodi, A.H.; Zaidan, B.B.; Zaidan, A.A.; Albahri, O.S.; Mohammed, K.I.; Malik, R.Q.; Almahdi, E.M.; Chyad, M.A.; Tareq, Z.; Albahri, A.S.; et al. Sentiment Analysis and Its Applications in Fighting COVID-19 and Infectious Diseases: A Systematic Review. Expert Syst. Appl. 2021 , 167 , 114155. [ Google Scholar ] [ CrossRef ]
- Jain, P.K.; Pamula, R.; Srivastava, G. A Systematic Literature Review on Machine Learning Applications for Consumer Sentiment Analysis Using Online Reviews. Comput. Sci. Rev. 2021 , 41 , 100413. [ Google Scholar ] [ CrossRef ]
- Rambocas, M.; Pacheco, B.G. Online Sentiment Analysis in Marketing Research: A Review. J. Res. Interact. Mark. 2018 , 12 , 146–163. [ Google Scholar ] [ CrossRef ]
- Zhang, X.; Guo, F.; Chen, T.; Pan, L.; Beliakov, G.; Wu, J. A Brief Survey of Machine Learning and Deep Learning Techniques for E-Commerce Research. J. Theor. Appl. Electron. Commer. Res. 2023 , 18 , 2188–2216. [ Google Scholar ] [ CrossRef ]
- Giachanou, A.; Crestani, F. Like It or Not: A Survey of Twitter Sentiment Analysis Methods. ACM Comput. Surv. 2016 , 49 , 1–41. [ Google Scholar ] [ CrossRef ]
- Krugmann, J.O.; Hartmann, J. Sentiment Analysis in the Age of Generative AI. Cust. Needs Solut. 2024 , 11 , 3. [ Google Scholar ] [ CrossRef ]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the Annual Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; pp. 4171–4186. [ Google Scholar ]
- Hartmann, J.; Heitmann, M.; Siebert, C.; Schamp, C. More than a Feeling: Accuracy and Application of Sentiment Analysis. Int. J. Res. Mark. 2023 , 40 , 75–87. [ Google Scholar ] [ CrossRef ]
- Wang, Z.; Xie, Q.; Feng, Y.; Ding, Z.; Yang, Z.; Xia, R. Is ChatGPT a Good Sentiment Analyzer? A Preliminary Study. arXiv 2023 , arXiv:2304.04339. [ Google Scholar ]
- Tsakalidis, A.; Papadopoulos, S.; Voskaki, R.; Ioannidou, K.; Boididou, C.; Cristea, A.I.; Liakata, M.; Kompatsiaris, Y. Building and Evaluating Resources for Sentiment Analysis in the Greek Language. Lang. Resour. Eval. 2018 , 52 , 1021–1044. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Bilianos, D. Experiments in Text Classification: Analyzing the Sentiment of Electronic Product Reviews in Greek. J. Quant. Linguist. 2022 , 29 , 374–386. [ Google Scholar ] [ CrossRef ]
- Markopoulos, G.; Mikros, G.; Iliadi, A.; Liontos, M. Sentiment Analysis of Hotel Reviews in Greek: A Comparison of Unigram Features. In Cultural Tourism in a Digital Era, Springer Proceedings in Business and Economics ; Katsoni, V., Ed.; Springer Science and Business Media B.V.: Berlin/Heidelberg, Germany, 2015; pp. 373–383. [ Google Scholar ]
- Dontaki, C.; Koukaras, P.; Tjortjis, C. Sentiment Analysis on English and Greek Twitter Data Regarding Vaccinations. In Proceedings of the 14th International Conference on Information, Intelligence, Systems and Applications, IISA, Volos, Greece, 10–12 July 2023; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2023. [ Google Scholar ]
- Charalampakis, B.; Spathis, D.; Kouslis, E.; Kermanidis, K. Detecting Irony on Greek Political Tweets: A Text Mining Approach. In Proceedings of the 16th International Conference on Engineering Applications of Neural Networks (INNS), Island, Rhodes, Greece, 25–28 September 2015; Association for Computing Machinery: New York, NY, USA, 2015. [ Google Scholar ]
- Athanasiou, V.; Maragoudakis, M. A Novel, Gradient Boosting Framework for Sentiment Analysis in Languages Where NLP Resources Are Not Plentiful: A Case Study for Modern Greek. Algorithms 2017 , 10 , 34. [ Google Scholar ] [ CrossRef ]
- Katika, A.; Zoulias, E.; Koufi, V.; Malamateniou, F. Mining Greek Tweets on Long COVID Using Sentiment Analysis and Topic Modeling. In Healthcare Transformation with Informatics and Artificial Intelligence ; IOS Press BV: Amsterdam, The Netherlands, 2023; Volume 305, pp. 545–548. [ Google Scholar ]
- Patsiouras, E.; Koroni, I.; Mademlis, I.; Pitas, I. GreekPolitics: Sentiment Analysis on Greek Politically Charged Tweets. In Proceedings of the 2023 31st European Signal Processing Conference (EUSIPCO), Helsinki, Finland, 4–8 September 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1320–1324. [ Google Scholar ]
- Alexandridis, G.; Varlamis, I.; Korovesis, K.; Caridakis, G.; Tsantilas, P. A Survey on Sentiment Analysis and Opinion Mining in Greek Social Media. Information 2021 , 12 , 331. [ Google Scholar ] [ CrossRef ]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed Representations of Words and Phrases and Their Compositionality. In Proceedings of the 26th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–8 December 2013; pp. 3111–3119. [ Google Scholar ]
- Pennington, J.; Socher, R.; Manning, C.D. GloVe: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [ Google Scholar ]
- Joulin, A.; Grave, E.; Bojanowski, P.; Mikolov, T. Bag of Tricks for Efficient Text Classification. In Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics, Valencia, Spain, 3–7 April 2017; pp. 427–431. [ Google Scholar ]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine Learning and Deep Learning. Electron. Mark. 2021 , 31 , 685–695. [ Google Scholar ] [ CrossRef ]
- Yadav, A.; Vishwakarma, D.K. Sentiment Analysis Using Deep Learning Architectures: A Review. Artif. Intell. Rev. 2020 , 53 , 4335–4385. [ Google Scholar ] [ CrossRef ]
- Vaswani, A.; Brain, G.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the NIPS’17: Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 6000–6010. [ Google Scholar ]
- Sanh, V.; Debut, L.; Chaumond, J.; Wolf, T. DistilBERT, a Distilled Version of BERT: Smaller, Faster, Cheaper and Lighter. arXiv 2019 , arXiv:1910.01108. [ Google Scholar ]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019 , arXiv:1907.11692. [ Google Scholar ]
- Koutsikakis, J.; Chalkidis, I.; Malakasiotis, P.; Androutsopoulos, I. GREEK-BERT: The Greeks Visiting Sesame Street. In Proceedings of the 11th Hellenic Conference on Artificial Intelligence, Athens, Greece, 2–4 September 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 110–117. [ Google Scholar ]
- Kalamatianos, G.; Mallis, D.; Symeonidis, S.; Arampatzis, A. Sentiment Analysis of Greek Tweets and Hashtags Using a Sentiment Lexicon. In Proceedings of the 19th Panhellenic Conference on Informatics, Athens, Greece, 1–3 October 2015; Association for Computing Machinery: New York, NY, USA, 2015; pp. 63–68. [ Google Scholar ]
- Kydros, D.; Argyropoulou, M.; Vrana, V. A Content and Sentiment Analysis of Greek Tweets during the Pandemic. Sustainability 2021 , 13 , 6150. [ Google Scholar ] [ CrossRef ]
- Samaras, L.; García-Barriocanal, E.; Sicilia, M.A. Sentiment Analysis of COVID-19 Cases in Greece Using Twitter Data. Expert. Syst. Appl. 2023 , 230 , 120577. [ Google Scholar ] [ CrossRef ]
- Giatsoglou, M.; Vozalis, M.G.; Diamantaras, K.; Vakali, A.; Sarigiannidis, G.; Chatzisavvas, K.C. Sentiment Analysis Leveraging Emotions and Word Embeddings. Expert. Syst. Appl. 2017 , 69 , 214–224. [ Google Scholar ] [ CrossRef ]
- Aivatoglou, G.; Fytili, A.; Arampatzis, G.; Zaikis, D.; Stylianou, N.; Vlahavas, I. End-to-End Aspect Extraction and Aspect-Based Sentiment Analysis Framework for Low-Resource Languages. In Lecture Notes in Networks and Systems, Proceedings of the Intelligent Systems and Applications, Amsterdam, The Netherlands, 7–8 September 2023 ; Springer: Cham, Switzerland, 2023; Volume 824, pp. 841–858. [ Google Scholar ]
- Fragkis, N. Skroutz Shops Greek Reviews. Available online: https://www.kaggle.com/datasets/nikosfragkis/skroutz-shop-reviews-sentiment-analysis (accessed on 13 April 2024).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011 , 12 , 2825–2830. [ Google Scholar ]
- Cauteruccio, F.; Kou, Y. Investigating the Emotional Experiences in ESports Spectatorship: The Case of League of Legends. Inf. Process Manag. 2023 , 60 , 103516. [ Google Scholar ] [ CrossRef ]
- Tamer, M.; Khamis, M.A.; Yahia, A.; Khaled, S.A.; Ashraf, A.; Gomaa, W. Arab Reactions towards Russo-Ukrainian War. EPJ Data Sci. 2023 , 12 , 36. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| ML Model | Training | Testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| LR | 92.73 | 92.77 | 92.73 | 92.73 | 93.52 | 93.59 | 93.52 | 93.51 |
| KNN | 81.43 | 83.11 | 81.43 | 81.22 | 80.93 | 83.22 | 80.93 | 80.55 |
| DT | 83.53 | 83.56 | 83.38 | 83.32 | 83.68 | 83.70 | 83.68 | 83.67 |
| MNB | ||||||||
| SVM | 90.63 | 90.67 | 90.63 | 90.63 | 91.61 | 91.69 | 91.61 | 91.60 |
| RF | 88.65 | 88.76 | 88.82 | 88.70 | 89.32 | 89.38 | 89.32 | 89.32 |
| AdaBoost | 89.35 | 89.43 | 89.35 | 89.35 | 90.77 | 90.83 | 90.77 | 90.76 |
| SGB | 89.43 | 89.48 | 89.47 | 89.45 | 89.47 | 89.48 | 89.47 | 89.47 |
| ML Model | Training | Testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| LR | 92.25 | 92.28 | 92.25 | 92.25 | 93.44 | 93.45 | 93.44 | 93.44 |
| KNN | 88.34 | 88.48 | 88.34 | 88.33 | 88.33 | 88.35 | 88.33 | 88.33 |
| DT | 82.79 | 82.80 | 82.29 | 82.77 | 82.07 | 82.08 | 82.07 | 82.07 |
| MNB | 92.37 | 92.72 | 92.37 | 92.35 | 93.06 | 93.35 | 93.06 | 93.05 |
| SVM | ||||||||
| RF | 88.74 | 88.88 | 88.84 | 88.92 | 89.09 | 89.10 | 89.09 | 89.09 |
| AdaBoost | 89.30 | 89.32 | 89.30 | 89.30 | 89.63 | 89.64 | 89.63 | 89.62 |
| SGB | 88.02 | 88.11 | 88.15 | 88.13 | 89.70 | 89.71 | 89.70 | 89.70 |
| ML Model | Training | Testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| LR | 92.75 | 92.79 | 92.75 | 92.75 | 93.44 | 93.52 | 93.44 | 93.44 |
| MNB | ||||||||
| SVM | 92.65 | 92.66 | 92.65 | 92.65 | 92.83 | 92.85 | 92.83 | 92.83 |
| ML Model | Training | Testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| LR | 93.49 | 93.51 | 93.49 | 93.49 | ||||
| MNB | 92.65 | 92.96 | 92.65 | 92.64 | 93.36 | 93.63 | 93.36 | 93.36 |
| SVM | 94.20 | 94.20 | 94.20 | 94.20 | ||||
| Model/ Neurons | Training | Testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| MLP/60 | 93.68 | 93.72 | 93.68 | 93.68 | 93.75 | 93.77 | 93.75 | 93.74 |
| MLP/70 | 93.78 | 93.80 | 93.78 | 93.78 | 94.58 | 94.59 | 94.58 | 94.58 |
| MLP/80 | 93.76 | 93.77 | 93.76 | 93.76 | 94.20 | 94.22 | 94.20 | 94.20 |
| MLP/90 | 93.90 | 93.93 | 93.90 | 93.90 | ||||
| MLP/100 | 93.82 | 93.83 | 93.82 | 93.82 | ||||
| Model/ Neurons | Training | Testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| MLP/60 | 93.30 | 93.33 | 93.30 | 93.30 | 94.51 | 94.51 | 94.51 | 94.51 |
| MLP/70 | ||||||||
| MLP/80 | 93.74 | 93.79 | 93.74 | 93.74 | 94.05 | 94.05 | 94.05 | 94.05 |
| MLP/90 | 93.78 | 93.81 | 93.78 | 93.78 | 94.51 | 94.51 | 94.51 | 94.51 |
| MLP/100 | 93.74 | 93.77 | 93.74 | 93.74 | 93.90 | 93.91 | 93.90 | 93.90 |
| Epochs | Training | Testing | ||
|---|---|---|---|---|
| Loss | Accuracy (%) | Loss | Accuracy (%) | |
| 1 | 0.26 | 89.66 | 0.15 | 94.74 |
| 2 | 0.11 | 96.01 | 0.12 | 95.42 |
| 3 | 0.08 | 97.25 | 0.12 | 95.88 |
| 4 | 0.05 | 98.24 | 0.13 | 96.03 |
| Model | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|
| GPT-3.5-turbo | 93.13 | 93.98 | 93.13 | 93.30 |
| GPT-4 |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Michailidis, P.D. A Comparative Study of Sentiment Classification Models for Greek Reviews. Big Data Cogn. Comput. 2024 , 8 , 107. https://doi.org/10.3390/bdcc8090107
Michailidis PD. A Comparative Study of Sentiment Classification Models for Greek Reviews. Big Data and Cognitive Computing . 2024; 8(9):107. https://doi.org/10.3390/bdcc8090107
Michailidis, Panagiotis D. 2024. "A Comparative Study of Sentiment Classification Models for Greek Reviews" Big Data and Cognitive Computing 8, no. 9: 107. https://doi.org/10.3390/bdcc8090107
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: 04 September 2024
CDK5–cyclin B1 regulates mitotic fidelity
- Xiao-Feng Zheng ORCID: orcid.org/0000-0001-8769-4604 1 na1 ,
- Aniruddha Sarkar ORCID: orcid.org/0000-0002-9393-1335 1 na1 ,
- Humphrey Lotana 2 ,
- Aleem Syed ORCID: orcid.org/0000-0001-7942-3900 1 ,
- Huy Nguyen ORCID: orcid.org/0000-0002-4424-1047 1 ,
- Richard G. Ivey 3 ,
- Jacob J. Kennedy 3 ,
- Jeffrey R. Whiteaker 3 ,
- Bartłomiej Tomasik ORCID: orcid.org/0000-0001-5648-345X 1 , 4 nAff7 ,
- Kaimeng Huang ORCID: orcid.org/0000-0002-0552-209X 1 , 5 ,
- Feng Li 1 ,
- Alan D. D’Andrea ORCID: orcid.org/0000-0001-6168-6294 1 , 5 ,
- Amanda G. Paulovich ORCID: orcid.org/0000-0001-6532-6499 3 ,
- Kavita Shah 2 ,
- Alexander Spektor ORCID: orcid.org/0000-0002-1085-3205 1 , 5 &
- Dipanjan Chowdhury ORCID: orcid.org/0000-0001-5645-3752 1 , 5 , 6
Nature ( 2024 ) Cite this article
Metrics details
CDK1 has been known to be the sole cyclin-dependent kinase (CDK) partner of cyclin B1 to drive mitotic progression 1 . Here we demonstrate that CDK5 is active during mitosis and is necessary for maintaining mitotic fidelity. CDK5 is an atypical CDK owing to its high expression in post-mitotic neurons and activation by non-cyclin proteins p35 and p39 2 . Here, using independent chemical genetic approaches, we specifically abrogated CDK5 activity during mitosis, and observed mitotic defects, nuclear atypia and substantial alterations in the mitotic phosphoproteome. Notably, cyclin B1 is a mitotic co-factor of CDK5. Computational modelling, comparison with experimentally derived structures of CDK–cyclin complexes and validation with mutational analysis indicate that CDK5–cyclin B1 can form a functional complex. Disruption of the CDK5–cyclin B1 complex phenocopies CDK5 abrogation in mitosis. Together, our results demonstrate that cyclin B1 partners with both CDK5 and CDK1, and CDK5–cyclin B1 functions as a canonical CDK–cyclin complex to ensure mitotic fidelity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
185,98 € per year
only 3,65 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others

Core control principles of the eukaryotic cell cycle
CDC7-independent G1/S transition revealed by targeted protein degradation
Evolution of opposing regulatory interactions underlies the emergence of eukaryotic cell cycle checkpoints
Data availability.
All data supporting the findings of this study are available in the Article and its Supplementary Information . The LC–MS/MS proteomics data have been deposited to the ProteomeXchange Consortium 60 via the PRIDE 61 partner repository under dataset identifier PXD038386 . Correspondence regarding experiments and requests for materials should be addressed to the corresponding authors.
Wieser, S. & Pines, J. The biochemistry of mitosis. Cold Spring Harb. Perspect. Biol. 7 , a015776 (2015).
Article PubMed PubMed Central Google Scholar
Dhavan, R. & Tsai, L. H. A decade of CDK5. Nat. Rev. Mol. Cell Biol. 2 , 749–759 (2001).
Article CAS PubMed Google Scholar
Malumbres, M. Cyclin-dependent kinases. Genome Biol. 15 , 122 (2014).
Coverley, D., Laman, H. & Laskey, R. A. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 4 , 523–528 (2002).
Desai, D., Wessling, H. C., Fisher, R. P. & Morgan, D. O. Effects of phosphorylation by CAK on cyclin binding by CDC2 and CDK2. Mol. Cell. Biol. 15 , 345–350 (1995).
Article CAS PubMed PubMed Central Google Scholar
Brown, N. R. et al. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 6 , 6769 (2015).
Article ADS CAS PubMed PubMed Central Google Scholar
Strauss, B. et al. Cyclin B1 is essential for mitosis in mouse embryos, and its nuclear export sets the time for mitosis. J. Cell Biol. 217 , 179–193 (2018).
Gavet, O. & Pines, J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189 , 247–259 (2010).
Barbiero, M. et al. Cell cycle-dependent binding between cyclin B1 and Cdk1 revealed by time-resolved fluorescence correlation spectroscopy. Open Biol. 12 , 220057 (2022).
Pines, J. & Hunter, T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58 , 833–846 (1989).
Clute, P. & Pines, J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1 , 82–87 (1999).
Potapova, T. A. et al. The reversibility of mitotic exit in vertebrate cells. Nature 440 , 954–958 (2006).
Basu, S., Greenwood, J., Jones, A. W. & Nurse, P. Core control principles of the eukaryotic cell cycle. Nature 607 , 381–386 (2022).
Santamaria, D. et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448 , 811–815 (2007).
Article ADS CAS PubMed Google Scholar
Zheng, X. F. et al. A mitotic CDK5-PP4 phospho-signaling cascade primes 53BP1 for DNA repair in G1. Nat. Commun. 10 , 4252 (2019).
Article ADS PubMed PubMed Central Google Scholar
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 13 , 397–406 (2014).
Article CAS Google Scholar
Pozo, K. & Bibb, J. A. The emerging role of Cdk5 in cancer. Trends Cancer 2 , 606–618 (2016).
Sharma, S. & Sicinski, P. A kinase of many talents: non-neuronal functions of CDK5 in development and disease. Open Biol. 10 , 190287 (2020).
Sun, K. H. et al. Novel genetic tools reveal Cdk5’s major role in Golgi fragmentation in Alzheimer’s disease. Mol. Biol. Cell 19 , 3052–3069 (2008).
Sharma, S. et al. Targeting the cyclin-dependent kinase 5 in metastatic melanoma. Proc. Natl Acad. Sci. USA 117 , 8001–8012 (2020).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14 , 431–441 (2018).
Simpson, L. M. et al. Target protein localization and its impact on PROTAC-mediated degradation. Cell Chem. Biol. 29 , 1482–1504 e1487 (2022).
Vassilev, L. T. et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl Acad. Sci. USA 103 , 10660–10665 (2006).
Janssen, A. F. J., Breusegem, S. Y. & Larrieu, D. Current methods and pipelines for image-based quantitation of nuclear shape and nuclear envelope abnormalities. Cells 11 , 347 (2022).
Thompson, S. L. & Compton, D. A. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl Acad. Sci. USA 108 , 17974–17978 (2011).
Kline-Smith, S. L. & Walczak, C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell 15 , 317–327 (2004).
Prosser, S. L. & Pelletier, L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18 , 187–201 (2017).
Zeng, X. et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18 , 382–395 (2010).
Warren, J. D., Orr, B. & Compton, D. A. A comparative analysis of methods to measure kinetochore-microtubule attachment stability. Methods Cell. Biol. 158 , 91–116 (2020).
Gregan, J., Polakova, S., Zhang, L., Tolic-Norrelykke, I. M. & Cimini, D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol 21 , 374–381 (2011).
Etemad, B., Kuijt, T. E. & Kops, G. J. Kinetochore-microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat. Commun. 6 , 8987 (2015).
Tauchman, E. C., Boehm, F. J. & DeLuca, J. G. Stable kinetochore-microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat. Commun. 6 , 10036 (2015).
Mitchison, T. & Kirschner, M. Microtubule assembly nucleated by isolated centrosomes. Nature 312 , 232–237 (1984).
Fourest-Lieuvin, A. et al. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol. Biol. Cell 17 , 1041–1050 (2006).
Ubersax, J. A. et al. Targets of the cyclin-dependent kinase Cdk1. Nature 425 , 859–864 (2003).
Yang, C. H., Lambie, E. J. & Snyder, M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 116 , 1303–1317 (1992).
Yang, C. H. & Snyder, M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell 3 , 1259–1267 (1992).
Kotak, S., Busso, C. & Gonczy, P. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 32 , 2517–2529 (2013).
Kitagawa, M. et al. Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Rep. 7 , 166–179 (2014).
Schrock, M. S. et al. MKLP2 functions in early mitosis to ensure proper chromosome congression. J. Cell Sci. 135 , jcs259560 (2022).
Sun, M. et al. NuMA regulates mitotic spindle assembly, structural dynamics and function via phase separation. Nat. Commun. 12 , 7157 (2021).
Chen, Q., Zhang, X., Jiang, Q., Clarke, P. R. & Zhang, C. Cyclin B1 is localized to unattached kinetochores and contributes to efficient microtubule attachment and proper chromosome alignment during mitosis. Cell Res. 18 , 268–280 (2008).
Kabeche, L. & Compton, D. A. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature 502 , 110–113 (2013).
Hegarat, N. et al. Cyclin A triggers mitosis either via the Greatwall kinase pathway or cyclin B. EMBO J. 39 , e104419 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596 , 583–589 (2021).
Wood, D. J. & Endicott, J. A. Structural insights into the functional diversity of the CDK-cyclin family. Open Biol. 8 , 180112 (2018).
Brown, N. R., Noble, M. E., Endicott, J. A. & Johnson, L. N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1 , 438–443 (1999).
Tarricone, C. et al. Structure and regulation of the CDK5-p25 nck5a complex. Mol. Cell 8 , 657–669 (2001).
Poon, R. Y., Lew, J. & Hunter, T. Identification of functional domains in the neuronal Cdk5 activator protein. J. Biol. Chem. 272 , 5703–5708 (1997).
Oppermann, F. S. et al. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteom. 8 , 1751–1764 (2009).
van den Heuvel, S. & Harlow, E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262 , 2050–2054 (1993).
Article ADS PubMed Google Scholar
Nakatani, Y. & Ogryzko, V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370 , 430–444 (2003).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11 , 2301–2319 (2016).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13 , 731–740 (2016).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 , e47 (2015).
R Core Team. R: a language and environment for statistical computing (2021).
Wickham, H. ggplot2: elegant graphics for data analysis (2016).
Slowikowski, K. ggrepel: automatically position non-overlapping text labels with “ggplot2” (2018).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2 , 100141 (2021).
CAS PubMed PubMed Central Google Scholar
Deutsch, E. W. et al. The ProteomeXchange consortium in 2020: enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 48 , D1145–D1152 (2020).
CAS PubMed Google Scholar
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 , D442–D450 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 , 139–140 (2010).
Nagahara, H. et al. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 4 , 1449–1452 (1998).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19 , 679–682 (2022).
Lu, C. et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 17 , 4291–4300 (2021).
Obenauer, J. C., Cantley, L. C. & Yaffe, M. B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31 , 3635–3641 (2003).
Download references
Acknowledgements
We thank D. Pellman for comments on the manuscript; W. Michowski, S. Sharma, P. Sicinski, B. Nabet and N. Gray for the reagents; J. A. Tainer for providing access to software used for structural analysis; and S. Gerber for sharing unpublished results. D.C. is supported by grants R01 CA208244 and R01 CA264900, DOD Ovarian Cancer Award W81XWH-15-0564/OC140632, Tina’s Wish Foundation, Detect Me If You Can, a V Foundation Award, a Gray Foundation grant and the Claudia Adams Barr Program in Innovative Basic Cancer Research. A. Spektor would like to acknowledge support from K08 CA208008, the Burroughs Wellcome Fund Career Award for Medical Scientists, Saverin Breast Cancer Research Fund and the Claudia Adams Barr Program in Innovative Basic Cancer Research. X.-F.Z. was an American Cancer Society Fellow and is supported by the Breast and Gynecologic Cancer Innovation Award from Susan F. Smith Center for Women’s Cancers at Dana-Farber Cancer Institute. A. Syed is supported by the Claudia Adams Barr Program in Innovative Basic Cancer Research. B.T. was supported by the Polish National Agency for Academic Exchange (grant PPN/WAL/2019/1/00018) and by the Foundation for Polish Science (START Program). A.D.D is supported by NIH grant R01 HL52725. A.G.P. by National Cancer Institute grants U01CA214114 and U01CA271407, as well as a donation from the Aven Foundation; J.R.W. by National Cancer Institute grant R50CA211499; and K.S. by NIH awards 1R01-CA237660 and 1RF1NS124779.
Author information
Bartłomiej Tomasik
Present address: Department of Oncology and Radiotherapy, Medical University of Gdańsk, Faculty of Medicine, Gdańsk, Poland
These authors contributed equally: Xiao-Feng Zheng, Aniruddha Sarkar
Authors and Affiliations
Division of Radiation and Genome Stability, Department of Radiation Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA
Xiao-Feng Zheng, Aniruddha Sarkar, Aleem Syed, Huy Nguyen, Bartłomiej Tomasik, Kaimeng Huang, Feng Li, Alan D. D’Andrea, Alexander Spektor & Dipanjan Chowdhury
Department of Chemistry and Purdue University Center for Cancer Research, Purdue University, West Lafayette, IN, USA
Humphrey Lotana & Kavita Shah
Translational Science and Therapeutics Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
Richard G. Ivey, Jacob J. Kennedy, Jeffrey R. Whiteaker & Amanda G. Paulovich
Department of Biostatistics and Translational Medicine, Medical University of Łódź, Łódź, Poland
Broad Institute of Harvard and MIT, Cambridge, MA, USA
Kaimeng Huang, Alan D. D’Andrea, Alexander Spektor & Dipanjan Chowdhury
Department of Biological Chemistry & Molecular Pharmacology, Harvard Medical School, Boston, MA, USA
Dipanjan Chowdhury
You can also search for this author in PubMed Google Scholar
Contributions
X.-F.Z., A. Sarkar., A. Spektor. and D.C. conceived the project and designed the experiments. X.-F.Z. and A. Sarkar performed the majority of experiments and associated analyses except as listed below. H.L. expressed relevant proteins and conducted the kinase activity assays for CDK5–cyclin B1, CDK5–p35 and CDK5(S46) variant complexes under the guidance of K.S.; A. Syed performed structural modelling and analysis. R.G.I., J.J.K. and J.R.W. performed MS and analysis. B.T. and H.N. performed MS data analyses. K.H. provided guidance to screen CDK5(as) knocked-in clones and performed sequence analysis to confirm CDK5(as) knock-in. F.L. and A.D.D. provided reagents and discussion on CDK5 substrates analyses. X.-F.Z., A. Sarkar, A. Spektor and D.C. wrote the manuscript with inputs and edits from all of the other authors.
Corresponding authors
Correspondence to Alexander Spektor or Dipanjan Chowdhury .
Ethics declarations
Competing interests.
A.D.D. reports consulting for AstraZeneca, Bayer AG, Blacksmith/Lightstone Ventures, Bristol Myers Squibb, Cyteir Therapeutics, EMD Serono, Impact Therapeutics, PrimeFour Therapeutics, Pfizer, Tango Therapeutics and Zentalis Pharmaceuticals/Zeno Management; is an advisory board member for Cyteir and Impact Therapeutics; a stockholder in Cedilla Therapeutics, Cyteir, Impact Therapeutics and PrimeFour Therapeutics; and reports receiving commercial research grants from Bristol Myers Squibb, EMD Serono, Moderna and Tango Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information.
Nature thanks Yibing Shan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 inhibition of cdk5 in analogue-sensitive (cdk5- as ) system..
a , Schematics depicting specific inhibition of the CDK5 analogue-sensitive ( as ) variant. Canonical ATP-analogue inhibitor (In, yellow) targets endogenous CDK5 (dark green) at its ATP-binding catalytic site nonspecifically since multiple kinases share structurally similar catalytic sites (left panel). The analogue-sensitive ( as , light green) phenylalanine-to-glycine (F80G) mutation confers a structural change adjacent to the catalytic site of CDK5 that does not impact its catalysis but accommodates the specific binding of a non-hydrolysable bulky orthogonal inhibitor 1NM-PP1(In*, orange). Introduction of 1NM-PP1 thus selectively inhibits CDK5- as variant (right panel). b , Immunoblots showing two clones (Cl 23 and Cl 50) of RPE-1 cells expressing FLAG-HA-CDK5- as in place of endogenous CDK5. Representative results are shown from three independent repeats. c , Proliferation curve of parental RPE-1 and RPE-1 CDK5- as cells. Data represent mean ± s.d. from three independent repeats. p -value was determined by Mann Whitney U test. d , Immunoblots showing immunoprecipitated CDK1-cyclin B1 complex or CDK5- as -cyclin B1 complex by the indicated antibody-coupled agarose, from nocodazole arrested RPE-1 CDK5- as cells with treated with or without 1NM-PP1 for inhibition of CDK5- as , from three independent replicate experiments. e , In-vitro kinase activity quantification of immunoprecipitated complex shown in d . Data represent mean ± s.d. from three independent experiments. p -values were determined by unpaired, two-tailed student’s t-test. f , Immunoblots of RPE-1 CDK5- as cells treated with either DMSO or 1NM-PP1 for 2 h prior to and upon release from RO-3306 and collected at 60 min following release. Cells were lysed and blotted with anti-bodies against indicated proteins (upper panel). Quantification of the relative intensity of PP4R3β phosphorylation at S840 in 1NM-PP1-treated CDK5- as cells compared to DMSO-treatment (lower panel). g , Experimental scheme for specific and temporal abrogation of CDK5 in RPE-1 CDK5- as cells. Data represent mean ± S.D from quadruplicate repeats. p -value was determined by one sample t and Wilcoxon test. h , Hoechst staining showing primary nuclei and micronuclei of RPE-1 CDK5- as with indicated treatment; scale bar is as indicated (left panel). Right, quantification of the percentage of cells with micronuclei after treatment. Data represent mean ± s.d. of three independent experiments from n = 2174 DMSO, n = 1788 1NM-PP1 where n is the number of cells. p- values were determined by unpaired, two-tailed student’s t-test. Scale bar is as indicated. Uncropped gel images are provided in Supplementary Fig. 1 .
Extended Data Fig. 2 Degradation of CDK5 in degradation tag (CDK5- dTAG ) system.
a , Schematic depicting the dTAG-13-inducible protein degradation system. Compound dTAG-13 links protein fused with FKBP12 F36V domain (dTAG) to CRBN-DDB1-CUL4A E3 ligase complex, leading to CRBN-mediated degradation. b , Immunoblots showing two clones of RPE-1 cells that express dTAG -HA-CDK5 in place of endogenous CDK5 (Cl N1 and Cl N4). Representative results are shown from three independent repeats. c , Proliferation curve of parental RPE-1 and RPE-1 CDK5-dTAG. Data represent mean ± s.d. of three independent repeats. p -value was determined by Mann Whitney U test. d and e , Representative images of RPE-1 CDK5- dTAG clone 1 (N1) ( d ) and RPE-1 CDK5- dTAG clone 4 (N4) ( e ) treated with DMSO or dTAG-13 for 2 h prior to and upon release from G2/M arrest and fixed at 120 min after release (top panel); quantification of CDK5 total intensity per cell (lower panels). Data represent mean ± s.d. of at least two independent experiments from n = 100 cells each condition. p- values were determined by unpaired, two-tailed student’s t-test. f , Immunoblots showing level of indicated proteins in RPE-1 CDK5- dTAG cells. Cells were treated with either DMSO or dTAG-13 for 2 h prior to and upon release from RO-3306 and lysed at 60 min following release (upper panel). Quantification of the relative intensity of PP4R3β phosphorylation at S840 in dTAG13-treated CDK5- dTAG cells compared to DMSO-treatment (lower panel). Data represent mean ± s.d. of four independent experiments. p -value was determined by one sample t and Wilcoxon test. g , Experimental scheme for specific and temporal abrogation of CDK5 in RPE-1 CDK5- dTAG cells. h , Hoechst staining showing primary nuclei and micronuclei of RPE-1 CDK5- dTAG with indicated treatment; scale bar is as indicated (left panel). Right, quantification of the percentage of cells with micronuclei after treatment. Data represent mean ± s.d. of three independent experiments from n = 2094 DMSO and n = 2095 dTAG-13, where n is the number of cells. p- values were determined by unpaired, two-tailed student’s t-test. Scale bar is as indicated. Uncropped gel images are provided in Supplementary Fig. 1 .
Extended Data Fig. 3 CDK5 abrogation render chromosome alignment and segregation defect despite intact spindle assembly checkpoint and timely mitotic duration.
a and b , Live-cell imaging snapshots of RPE-1 CDK5- as cells ( a ) and RPE-1 CDK5- dTAG cells ( b ) expressing mCherry-H2B and GFP-α-tubulin, abrogated of CDK5 by treatment with 1NM-PP1 or dTAG-13, respectively. Imaging commenced in prophase following release from RO-3306 into fresh media containing indicated chemicals (left); quantification of the percentage of cells with abnormal nuclear morphology (right). c and d , Representative snapshots of the final frame prior to metaphase-to-anaphase transition from a live-cell imaging experiment detailing chromosome alignment at the metaphase plate of RPE- CDK5- as (c) and RPE-1 CDK5- dTAG ( d ) expressing mCherry-H2B, and GFP-α-tubulin (left); quantification of the percentage of cells displaying abnormal chromosome alignment following indicated treatments (top right). e , Representative images showing the range of depolymerization outcomes (low polymers, high polymers and spindle-like) in DMSO- and 1NM-PP1-treated cells, as shown in Fig. 2e , from n = 50 for each condition, where n is number of metaphase cells . f , Quantifications of mitotic duration from nuclear envelope breakdown (NEBD) to anaphase onset of RPE-1 CDK5- as (left ) and RPE-1 CDK5- dTAG (right) cells, following the indicated treatments. Live-cell imaging of RPE-1 CDK5- as and RPE-1 CDK5- dTAG cells expressing mCherry-H2B and GFP-BAF commenced following release from RO-3306 arrest into fresh media containing DMSO or 1NM-PP1 or dTAG-13. g , Quantifications of the percentage of RPE-1 CDK5- as (left) and RPE-1 CDK5- dTAG (right) cells that were arrested in mitosis following the indicated treatments. Imaging commenced in prophase cells as described in a , following release from RO-3306 into fresh media in the presence or absence nocodazole as indicated. The data in a, c , and g represent mean ± s.d. of at least two independent experiments from n = 85 DMSO and n = 78 1NM-PP1 in a and c ; from n = 40 cells for each treatment condition in g . The data in b , d , and f represent mean ± s.d. of three independent experiments from n = 57 DMSO and n = 64 dTAG-13 in b and d ; from n = 78 DMSO and n = 64 1NM-PP1; n = 59 DMSO and n = 60 dTAG-13, in f , where n is the number of cells. p- values were determined by unpaired, two-tailed student’s t-test. Scale bar is as indicated.
Extended Data Fig. 4 CDK5 and CDK1 regulate tubulin dynamics.
a, b , Immunostaining of RPE-1 cells with antibodies against CDK1 and α-tubulin ( a ); and CDK5 and α-tubulin ( b ) at indicated stages of mitosis. c, d , Manders’ overlap coefficient M1 (CDK1 versus CDK5 on α-tubulin) ( c ); and M2 (α-tubulin on CDK1 versus CDK5) ( d ) at indicated phases of mitosis in cells shown in a and b . The data represent mean ± s.d. of at least two independent experiments from n = 25 cells in each mitotic stage. p- values were determined by unpaired, two-tailed student’s t-test.
Extended Data Fig. 5 Phosphoprotoemics analysis to identify mitotic CDK5 substrates.
a , Scheme of cell synchronization for phosphoproteomics: RPE-1 CDK5- as cells were arrested at G2/M by treatment with RO-3306 for 16 h. The cells were treated with 1NM-PP1 to initiate CDK5 inhibition. 2 h post-treatment, cells were released from G2/M arrest into fresh media with or without 1NM-PP1 to proceed through mitosis with or without continuing inhibition of CDK5. Cells were collected at 60 min post-release from RO-3306 for lysis. b , Schematic for phosphoproteomics-based identification of putative CDK5 substrates. c , Gene ontology analysis of proteins harbouring CDK5 inhibition-induced up-regulated phosphosites. d , Table indicating phospho-site of proteins that are down-regulated as result of CDK5 inhibition. e , Table indicating the likely kinases to phosphorylate the indicated phosphosites of the protein, as predicted by Scansite 4 66 . Divergent score denotes the extent by which phosphosite diverge from known kinase substrate recognition motif, hence higher divergent score indicating the corresponding kinase is less likely the kinase to phosphorylate the phosphosite.
Extended Data Fig. 6 Cyclin B1 is a mitotic co-factor of CDK5 and of CDK1.
a , Endogenous CDK5 was immunoprecipitated from RPE-1 cells collected at time points corresponding to the indicated cell cycle stage. Cell lysate input and elution of immunoprecipitation were immunoblotted by antibodies against the indicated proteins. RPE-1 cells were synchronized to G2 by RO-3306 treatment for 16 h and to prometaphase (M) by nocodazole treatment for 6 h. Asynch: Asynchronous. Uncropped gel images are provided in Supplementary Fig. 1 . b , Immunostaining of RPE-1 cells with antibodies against the indicated proteins at indicated mitotic stages (upper panels). Manders’ overlap coefficient M1 (Cyclin B1 on CDK1) and M2 (CDK1 on Cyclin B1) at indicated mitotic stages for in cells shown in b (lower panels). The data represent mean ± s.d. of at least two independent experiments from n = 25 mitotic cells in each mitotic stage. p- values were determined by unpaired, two-tailed student’s t-test. c , Table listing common proteins as putative targets of CDK5, uncovered from the phosphoproteomics anlaysis of down-regulated phosphoproteins upon CDK5 inhibition (Fig. 3 and Supplementary Table 1 ), and those of cyclin B1, uncovered from phosphoproteomics analysis of down-regulated phospho-proteins upon cyclin B1 degradation (Fig. 6 and Table EV2 in Hegarat et al. EMBO J. 2020). Proteins relevant to mitotic functions are highlighted in red.
Extended Data Fig. 7 Structural prediction and analyses of the CDK5-cyclin B1 complex.
a , Predicted alignment error (PAE) plots of the top five AlphaFold2 (AF2)-predicted models of CDK5-cyclin B1 (top row) and CDK1-cyclin B1 (bottom row) complexes, ranked by interface-predicted template (iPTM) scores. b , AlphaFold2-Multimer-predicted structure of the CDK5-cyclin B1 complex. c , Structural comparison of CDK-cyclin complexes. Left most panel: Structural-overlay of AF2 model of CDK5-cyclin B1 and crystal structure of phospho-CDK2-cyclin A3-substrate complex (PDB ID: 1QMZ ). The zoomed-in view of the activation loops of CDK5 and CDK2 is shown in the inset. V163 (in CDK5), V164 (in CDK2) and Proline at +1 position in the substrates are indicated with arrows. Middle panel: Structural-overlay of AF2 model of CDK5-cyclin B1 and crystal structure of CDK1-cyclin B1-Cks2 complex (PDB ID: 4YC3 ). The zoomed-in view of the activation loops of CDK5 and CDK1 is shown in the inset. Cks2 has been removed from the structure for clarity. Right most panel: structural-overlay of AF2 models of CDK5-cyclin B1 and CDK1-cyclin B1 complex. The zoomed view of the activation loops of CDK5 and CDK1 is shown in the inset. d , Secondary structure elements of CDK5, cyclin B1 and p25. The protein sequences, labelled based on the structural models, are generated by PSPript for CDK5 (AF2 model) ( i ), cyclin B1 (AF2 model) ( ii ) and p25 (PDB ID: 3O0G ) ( iii ). Structural elements ( α , β , η ) are defined by default settings in the program. Key loops highlighted in Fig. 4d are mapped onto the corresponding sequence.
Extended Data Fig. 8 Phosphorylation of CDK5 S159 is required for kinase activity and mitotic fidelity.
a , Structure of the CDK5-p25 complex (PDB ID: 1h41 ). CDK5 (blue) interacts with p25 (yellow). Serine 159 (S159, magenta) is in the T-loop. b , Sequence alignment of CDK5 and CDK1 shows that S159 in CDK5 is the analogous phosphosite as that of T161 in CDK1 for T-loop activation. Sequence alignment was performed by CLC Sequence Viewer ( https://www.qiagenbioinformatics.com/products/clc-sequence-viewer/ ). c , Immunoblots of indicated proteins in nocodazole-arrested mitotic (M) and asynchronous (Asy) HeLa cell lysate. d , Myc-His-tagged CDK5 S159 variants expressed in RPE-1 CDK5- as cells were immunoprecipitated from nocodazole-arrested mitotic lysate by Myc-agarose. Input from cell lysate and elution from immunoprecipitation were immunoblotted with antibodies against indicated protein. EV= empty vector. In vitro kinase activity assay of the indicated immunoprecipitated complex shown on the right panel. Data represent mean ± s.d. of four independent experiments. p -values were determined by unpaired two-tailed student’s t-test. e , Immunoblots showing RPE-1 FLAG-CDK5- as cells stably expressing Myc-His-tagged CDK5 WT and S159A, which were used in live-cell imaging and immunofluorescence experiments to characterize chromosome alignment and spindle architecture during mitosis, following inhibition of CDK5- as by 1NM-PP1, such that only the Myc-His-tagged CDK5 WT and S159A are not inhibited. Representative results are shown from three independent repeats. f , Hoechst staining showing nuclear morphology of RPE-1 CDK5- as cells expressing indicated CDK5 S159 variants following treatment with either DMSO or 1NMP-PP1 and fixation at 120 min post-release from RO-3306-induced arrest (upper panel); quantification of nuclear circularity and solidity (lower panels) g , Snapshots of live-cell imaging RPE-1 CDK5- as cells expressing indicated CDK5 S159 variant, mCherry-H2B, and GFP-α-tubulin, after release from RO-3306-induced arrest at G2/M, treated with 1NM-PP1 2 h prior to and upon after release from G2/M arrest (upper panel); quantification of cells displaying abnormal chromosome alignment in (lower panel). Representative images are shown from two independent experiments, n = 30 cells each cell line. h , Representative images of RPE-1 CDK5- as cells expressing indicated CDK5 S159 variants in metaphase, treated with DMSO or 1NM-PP1 for 2 h prior to and upon release from RO-3306-induced arrest, and then released into media containing 20 µM proTAME for 2 h, fixed and stained with tubulin and DAPI (upper panel); metaphase plate width and spindle length measurements for these representative cells were shown in the table on right; quantification of metaphase plate width and spindle length following the indicated treatments (lower panel). Data in f and h represent mean ± s.d. of at least two independent experiments from n = 486 WT, n = 561 S159A, and n = 401 EV, where n is the number of cells in f ; from n = 65 WT, n = 64 S159A, and n = 67 EV, where n is the number of cells in h . Scale bar is as indicated. Uncropped gel images are provided in Supplementary Fig. 1 .
Extended Data Fig. 9 The CDK5 co-factor-binding helix regulates CDK5 kinase activity.
a , Structure of the CDK5-p25 complex (PDB ID: 1h41 ). CDK5 (blue) interacts with p25 (yellow) at the PSSALRE helix (green). Serine 46 (S46, red) is in the PSSALRE helix. Serine 159 (S159, magenta) is in the T-loop. b , Sequence alignment of CDK5 and CDK1 shows that S46 is conserved in CDK1 and CDK5. Sequence alignment was performed by CLC Sequence Viewer ( https://www.qiagenbioinformatics.com/products/clc-sequence-viewer/ ). c , Immunoblots of CDK5 immunoprecipitation from lysate of E. coli BL21 (DE3) expressing His-tagged human CDK5 WT or CDK5 S46D, mixed with lysate of E. coli BL21 (DE3) expressing His-tagged human cyclin B1. Immunoprecipitated CDK5 alone or in the indicated complex were used in kinase activity assay, shown in Fig. 5b . Representative results are shown from three independent repeats. d , Immunoblots showing RPE-1 FLAG-CDK5- as cells stably expressing Myc-His-tagged CDK5 S46 phospho-variants, which were used in live-cell imaging and immunofluorescence experiments to characterize chromosome alignment and spindle architecture during mitosis, following inhibition of CDK5- as by 1NM-PP1, such that only the Myc-His-tagged CDK5 S46 phospho-variants are not inhibited. Representative results are shown from three independent repeats. e , Immunostaining of RPE-1 CDK5- as cells expressing Myc-His-tagged CDK5 WT or S46D with anti-PP4R3β S840 (pS840) antibody following indicated treatment (DMSO vs 1NM-PP1). Scale bar is as indicated (left). Normalized intensity level of PP4R3β S840 phosphorylation (right). Data represent mean ± s.d. of at least two independent experiments from n = 40 WT and n = 55 S46D, where n is the number of metaphase cells. p- values were determined by unpaired two-tailed student’s t-test. f , Immunoblots showing level of indicated proteins in RPE-1 CDK5- as cells expressing Myc-His-tagged CDK5 WT or S46D. Cells were treated with either DMSO or 1NM-PP1 for 2 h prior to and upon release from RO-3306 and collected and lysed at 60 min following release (left). Quantification of the intensity of PP4R3β phosphorylation at S840 (right). Data represent mean ± s.d. of four independent experiments. p -values were determined by two-tailed one sample t and Wilcoxon test. g , Representative snapshots of live-cell imaging of RPE-1 CDK5- as cells harbouring indicated CDK5 S46 variants expressing mCherry-H2B and GFP-α-tubulin, treated with 1NM-PP1, as shown in Fig. 5d , from n = 35 cells. Imaging commenced in prophase following release from RO-3306 into fresh media containing indicated chemicals. Uncropped gel images are provided in Supplementary Fig. 1 .
Extended Data Fig. 10 Localization of CDK5 S46 phospho-variants.
Immunostaining of RPE-1 CDK5- as cells stably expressing Myc-His CDK5-WT ( a ), S46A ( b ), and S46D ( c ) with antibodies against indicated protein in prophase, prometaphase, and metaphase. Data represent at least two independent experiments from n = 25 cells of each condition in each mitotic stage.
Extended Data Fig. 11 RPE-1 harbouring CDK5- as introduced by CRISPR-mediated knock-in recapitulates chromosome mis-segregation defects observed in RPE-1 overexpressing CDK5- as upon inhibition of CDK5- as by 1NM-PP1 treatment.
a , Chromatogram showing RPE-1 that harbours the homozygous CDK5- as mutation F80G introduced by CRISPR-mediated knock-in (lower panel), replacing endogenous WT CDK5 (upper panel). b , Immunoblots showing level of CDK5 expressed in parental RPE-1 and RPE-1 that harbours CDK5- as F80G mutation in place of endogenous CDK5. c , Representative images of CDK5- as knocked-in RPE-1 cells exhibiting lagging chromosomes following indicated treatments. d , Quantification of percentage of cells exhibiting lagging chromosomes following indicated treatments shown in (c). Data represent mean ± s.d. of three independent experiments from n = 252 DMSO, n = 220 1NM-PP1, where n is the number of cells. p -value was determined by two-tailed Mann Whitney U test.
Extended Data Fig. 12 CDK5 is highly expressed in post-mitotic neurons and overexpressed in cancers.
a , CDK5 RNAseq expression in tumours (left) with matched normal tissues (right). The data are analysed using 22 TCGA projects. Note that CDK5 expression is higher in many cancers compared to the matched normal tissues. BLCA, urothelial bladder carcinoma; BRCA, breast invasive carcinoma; CESC cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma; and UCEC, uterine corpus endometrial carcinoma. p -value was determined by two-sided Student’s t-test. ****: p <= 0.0001; ***: p <= 0.001; **: p <= 0.01; *: p <= 0.05; ns: not significant, p > 0.05. b , Scatter plots showing cells of indicated cancer types that are more dependent on CDK5 and less dependent on CDK1. Each dot represents a cancer cell line. The RNAi dependency data (in DEMETER2) for CDK5 and CDK1 were obtained from the Dependency Map ( depmap.org ). The slope line represents a simple linear regression analysis for the indicated cancer type. The four indicated cancer types (Head/Neck, Ovary, CNS/Brain, and Bowel) showed a trend of more negative CDK5 RNAi effect scores (indicative of more dependency) with increasing CDK1 RNAi effect scores (indicative of less dependency). The p value represents the significance of the correlation computed from a simple linear regression analysis of the data. Red circle highlights the subset of the cells that are relatively less dependent on CDK1 but more dependent on CDK5. c , Scatter plots showing bowel cancer cells that expresses CDK5 while being less dependent on CDK1. Each dot represents a cancer cell line. The data on gene effect of CDK1 CRISPR and CDK5 mRNA level were obtained from the Dependency Map ( depmap.org ). The slope line represents a simple linear regression analysis. Red circle highlights the subset of cells that are relatively less dependent on CDK1 but expresses higher level of CDK5. For b and c , solid line represents the best-fit line from simple linear regression using GraphPad Prism. Dashed lines represent 95% confidence bands of the best-fit line. p -value is determined by the F test testing the null hypothesis that the slope is zero. d , Scatter plots showing rapidly dividing cells of indicated cancer types that are more dependent on CDK5 and less dependent on CDK1. Each dots represents a cancer cell line. The doubling time data on the x-axis were obtained from the Cell Model Passports ( cellmodelpassports.sanger.ac.uk ). The RNAi dependency data (in DEMETER2) for CDK5, or CDK1, on the y-axis were obtained from the Dependency Map ( depmap.org ). Only cell lines with doubling time of less than 72 h are displayed and included for analysis. Each slope line represents a simple linear regression analysis for each cancer type. The indicated three cancer types were analysed and displayed because they showed a trend of faster proliferation rate (lower doubling time) with more negative CDK5 RNAi effect (more dependency) but increasing CDK1 RNAi effect (less dependency) scores. The p value represents the significance of the association of the three cancer types combined, computed from a multiple linear regression analysis of the combined data, using cancer type as a covariate. Red circle depicts subset of fast dividing cells that are relatively more dependent on CDK5 (left) and less dependent on CDK1 (right). Solid lines represent the best-fit lines from individual simple linear regressions using GraphPad Prism. p -value is for the test with the null hypothesis that the effect of the doubling time is zero from the multiple linear regression RNAi ~ Intercept + Doubling Time (hours) + Lineage.
Supplementary information
Supplementary figure 1.
Full scanned images of all western blots.
Reporting Summary
Peer review file, supplementary table 1.
Phosphosite changes in 1NM-PP1-treated cells versus DMSO-treated controls as measured by LC–MS/MS.
Supplementary Table 2
Global protein changes in 1NM-PP1-treated cells versus DMSO-treated controls as measured by LC–MS/MS.
Supplementary Video 1
RPE-1 CDK5(as) cell after DMSO treatment, ×100 imaging.
Supplementary Video 2
RPE-1 CDK5(as) cell after 1NM-PP1 treatment (example 1), ×100 imaging.
Supplementary Video 3
RPE-1 CDK5(as) cell after 1NM-PP1 treatment (example 2), ×100 imaging.
Supplementary Video 4
RPE-1 CDK5(dTAG) cell after DMSO treatment, ×100 imaging.
Supplementary Video 5
RPE-1 CDK5(dTAG) cell after dTAG-13 treatment (example 1), ×100 imaging.
Supplementary Video 6
RPE-1 CDK5(dTAG) cell after dTAG-13 treatment (example 2) ×100 imaging.
Supplementary Video 7
RPE-1 CDK5(as) cells expressing MYC-CDK5(WT) after 1NM-PP1 treatment, ×20 imaging.
Supplementary Video 8
RPE-1 CDK5(as) cells expressing MYC-EV after 1NM-PP1 treatment, ×20 imaging.
Supplementary Video 9
RPE-1 CDK5(as) cells expressing MYC-CDK5(S159A) after 1NM-PP1 treatment (example 1), ×20 imaging.
Supplementary Video 10
RPE-1 CDK5(as) cells expressing MYC-CDK5(S159A) after 1NM-PP1 treatment (example 2), ×20 imaging.
Supplementary Video 11
RPE-1 CDK5(as) cells expressing MYC-CDK5(WT) after 1NM-PP1 treatment, ×100 imaging.
Supplementary Video 12
RPE-1 CDK5(as) cells expressing MYC-CDK5(S46A) after 1NM-PP1 treatment (example 1), ×100 imaging.
Supplementary Video 13
RPE-1 CDK5(as) cells expressing MYC-CDK5(S46A) after 1NM-PP1 treatment (example 2), ×100 imaging.
Supplementary Video 14
RPE-1 CDK5(as) cells expressing MYC-CDK5(S46D) after 1NM-PP1 treatment (example 1), ×100 imaging.
Supplementary Video 15
RPE-1 CDK5(as) cells expressing MYC-CDK5(S46D) after 1NM-PP1 treatment (example 2), ×100 imaging.
Supplementary Video 16
RPE-1 CDK5(as) cells expressing MYC-EV after 1NM-PP1 treatment,×100 imaging.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Cite this article.
Zheng, XF., Sarkar, A., Lotana, H. et al. CDK5–cyclin B1 regulates mitotic fidelity. Nature (2024). https://doi.org/10.1038/s41586-024-07888-x
Download citation
Received : 24 March 2023
Accepted : 30 July 2024
Published : 04 September 2024
DOI : https://doi.org/10.1038/s41586-024-07888-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

COMMENTS
In this post, you will discover a cheat sheet for the most popular statistical hypothesis tests for a machine learning project with examples using the Python API. Each statistical test is presented in a consistent way, including: The name of the test. What the test is checking. The key assumptions of the test. How the test result is interpreted.
A hypothesis test is a formal statistical test we use to reject or fail to reject some statistical hypothesis. This tutorial explains how to perform the following hypothesis tests in Python: One sample t-test; Two sample t-test; Paired samples t-test; Let's jump in! Example 1: One Sample t-test in Python
It tests the null hypothesis that the population variances are equal (called homogeneity of variance or homoscedasticity). Suppose the resulting p-value of Levene's test is less than the significance level (typically 0.05).In that case, the obtained differences in sample variances are unlikely to have occurred based on random sampling from a population with equal variances.
Welcome to Hypothesis! Hypothesis is a Python library for creating unit tests which are simpler to write and more powerful when run, finding edge cases in your code you wouldn't have thought to look for. It is stable, powerful and easy to add to any existing test suite. It works by letting you write tests that assert that something should be ...
In this article, we interactively explore and visualize the difference between three common statistical tests: T-test, ANOVA test and Chi-Squared test. We also use examples to walkthrough essential steps in hypothesis testing: 1. define the null and alternative hypothesis. 2. choose the appropriate test.
Dive into the fascinating process of hypothesis testing with Python in this comprehensive guide. Perfect for aspiring data scientists and analytical minds, learn how to validate your predictions using statistical tests and Python's robust libraries. From understanding the basics of hypothesis formulation to executing detailed statistical analysis, this article illuminates the path to data ...
The process of hypothesis testing involves four steps: Now that we have a basic understanding of the concept, let's move on to the implementation in Python. We will use the scipy library to ...
Hypothesis testing is used to address questions about a population based on a subset from that population. For example, A/B testing is a framework for learning about consumer behavior based on a small sample of consumers. This course assumes some preexisting knowledge of Python, including the NumPy and pandas libraries.
Master hypothesis testing with Python: Learn statistical validation and data-driven decision-making in a concise guide. Boost your analysis skills with essential insights and resources.
Hypothesis testing uses statistics to test whether anything statistically significant has changed or not i.e. are our two distributions different because of luck or are they really different? A null hypothesis (𝐻0) is put forward which states that nothing has changed and an alternative hypothesis (𝐻1 or 𝐻𝐴) is proposed indicating ...
Statistical hypothesis testing allows researchers to make inferences about populations based on sample data. It involves setting up a null hypothesis, choosing a confidence level, calculating a p-value, and conducting tests such as two-tailed, one-tailed, or paired sample tests to draw conclusions.
Hypothesis testing is the act of testing whether a hypothesis or inference is true. When an alternate hypothesis is introduced, we test it against the null hypothesis to know which is correct. ... Numpy is a Python library used for scientific computing. It has a large library of functions for working with arrays. Scipy is a library for ...
Hypothesis testing lets you answer questions about your datasets in a statistically rigorous way. In this course, you'll grow your Python analytical skills as you learn how and when to use common tests like t-tests, proportion tests, and chi-square tests. Working with real-world data, including Stack Overflow user feedback and supply-chain data ...
Python is an open-source programming language that provides a Hypothesis library for property-based testing. Hypothesis testing in Python provides a framework for generating diverse and random test data, allowing development and testing teams to thoroughly test their code against a broad spectrum of inputs.
A hypothesis test verifies the credibility of the hypothesis and the likelihood that a finding observed from the sample data occurred by chance. We can use Python programming language to test ...
To use Hypothesis in this example, we import the given, strategies and assume in-built methods.. The @given decorator is placed just before each test followed by a strategy.. A strategy is specified using the strategy.X method which can be st.list(), st.integers(), st.text() and so on.. Here's a comprehensive list of strategies.. Strategies are used to generate test data and can be heavily ...
Permutation sampling is a great way to simulate the hypothesis that two variables have identical probability distributions. This is often a hypothesis you want to test, so in this exercise, you will write a function to generate a permutation sample from two data sets. Remember, a permutation sample of two arrays having respectively n1 and n2 ...
In this article, we are going to examine a case study of hypothesis testing on the seeds dataset, by using the Pingouin Python library. The Basic Steps of Hypothesis Testing. The first step in hypothesis testing is coming up with the research hypothesis, a statement that can be tested statistically and involves the comparison of variables, e.g ...
Hypothesis Testing in Python. In this course, you'll learn advanced statistical concepts like significance testing and multi-category chi-square testing, which will help you perform more powerful and robust data analysis. Enroll for free. Part of the Data Analyst (Python), and Data Scientist (Python) paths. 4.8 (359 reviews)
A detail: This works because Hypothesis ignores any arguments it hasn't been told to provide (positional arguments start from the right), so the self argument to the test is simply ignored and works as normal. This also means that Hypothesis will play nicely with other ways of parameterizing tests. e.g it works fine if you use pytest fixtures ...
A hypothesis is a claim or a premise that we want to test. Hypothesis testing is a way of backing up your conclusions with data, in a more "scientific" way. It is useful not only to scientists, but actually important in so many areas, ranging from marketing to web design to pharmaceutical trials and much more.
Our null hypothesis here is "Innocent" while the alternate is "Guilty". There are four outcomes possible now: Decided Guilty, Actual Guilty. Decided Innocent, Actual Innocent. Decided ...
In recent years, people have expressed their opinions and sentiments about products, services, and other issues on social media platforms and review websites. These sentiments are typically classified as either positive or negative based on their text content. Research interest in sentiment analysis for text reviews written in Greek is limited compared to that in English.
p-value is determined by the F test testing the null hypothesis that the slope is zero. d , Scatter plots showing rapidly dividing cells of indicated cancer types that are more dependent on CDK5 ...
Using Hypothesis settings for property-based testing of Python code Upping your game: Using composite strategies. So far, the examples I've used are simple. Hypothesis can handle much more complex test cases using composite strategies, which, as the name suggests, allows you to combine strategies to generate testing examples.